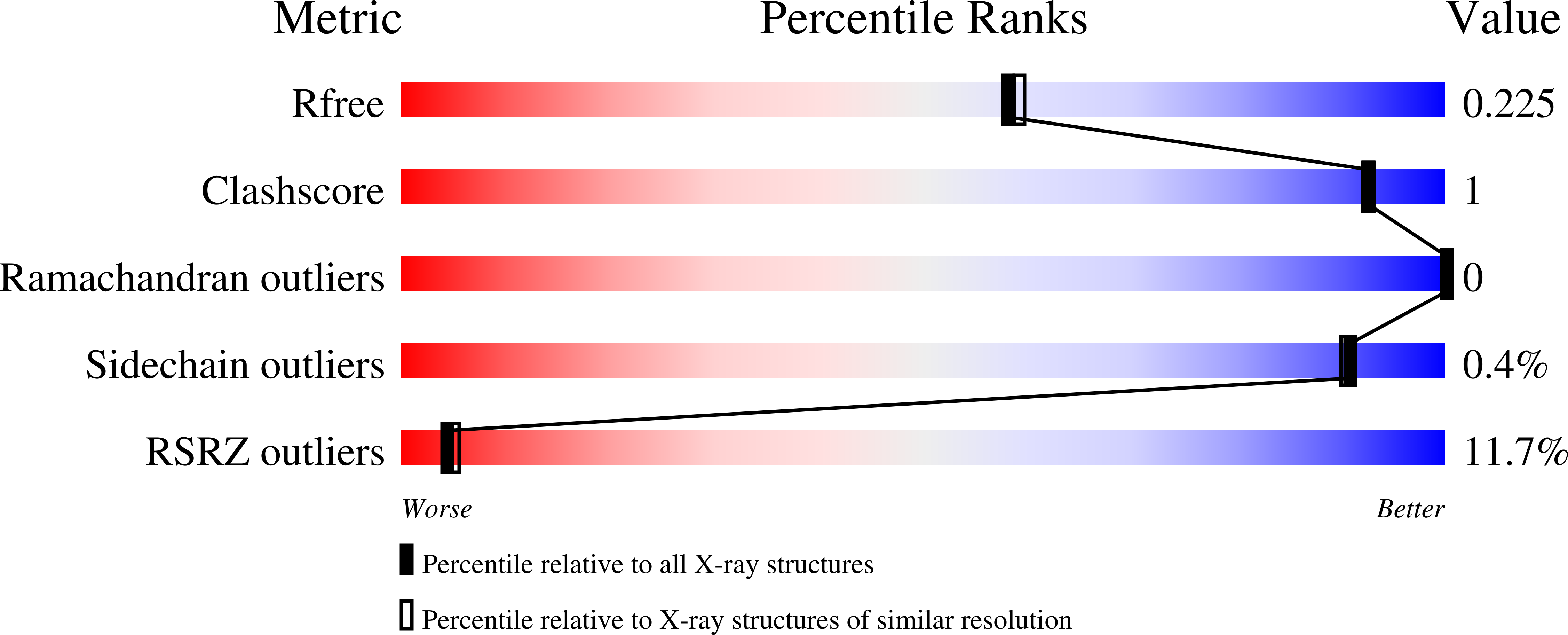
1 alpha,20S-Dihydroxyvitamin D3 Interacts with Vitamin D Receptor: Crystal Structure and Route of Chemical Synthesis.
Lin, Z., Chen, H., Belorusova, A.Y., Bollinger, J.C., Tang, E.K.Y., Janjetovic, Z., Kim, T.K., Wu, Z., Miller, D.D., Slominski, A.T., Postlethwaite, A.E., Tuckey, R.C., Rochel, N., Li, W.(2017) Sci Rep 7: 10193-10193
- PubMed: 28860545
- DOI: https://doi.org/10.1038/s41598-017-10917-7
- Primary Citation of Related Structures:
5MX7 - PubMed Abstract:
1α,20S-Dihydroxyvitamin D3 [1,20S(OH) 2 D 3 ], a natural and bioactive vitamin D3 metabolite, was chemically synthesized for the first time. X-ray crystallography analysis of intermediate 15 confirmed its 1α-OH configuration. 1,20S(OH) 2 D 3 interacts with the vitamin D receptor (VDR), with similar potency to its native ligand, 1α,25-dihydroxyvitamin D 3 [1,25(OH) 2 D 3 ] as illustrated by its ability to stimulate translocation of the VDR to the nucleus, stimulate VDRE-reporter activity, regulate VDR downstream genes (VDR, CYP24A1, TRPV6 and CYP27B1), and inhibit the production of inflammatory markers (IFNγ and IL1β). However, their co-crystal structures revealed differential molecular interactions of the 20S-OH moiety and the 25-OH moiety to the VDR, which may explain some differences in their biological activities. Furthermore, this study provides a synthetic route for the synthesis of 1,20S(OH) 2 D 3 using the intermediate 1α,3β-diacetoxypregn-5-en-20-one (3), and provides a molecular and biological basis for the development of 1,20S(OH) 2 D 3 and its analogs as potential therapeutic agents.
Organizational Affiliation:
Department of Pharmaceutical Sciences, University of Tennessee Health Science Center, 881 Madison Avenue, Room 561, Memphis, TN, 38163, United States.