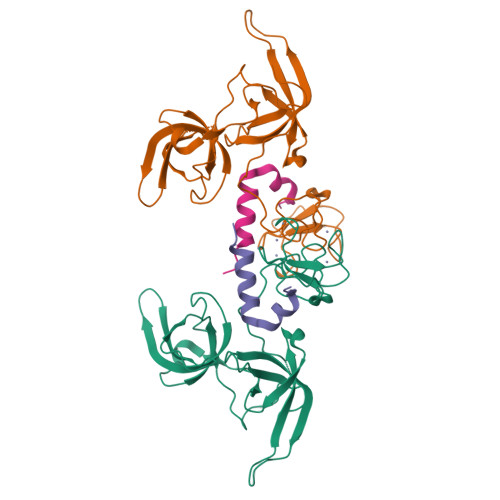
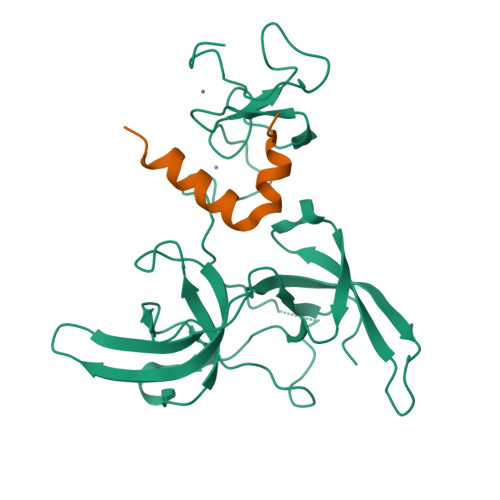
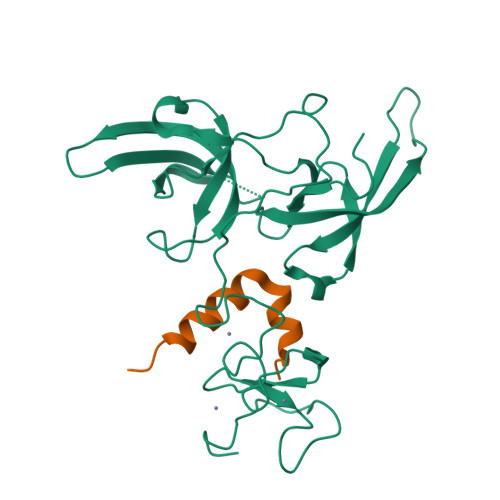
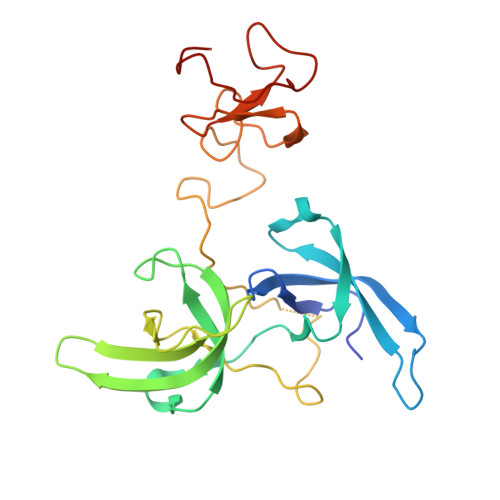
Defining ortholog-specific UHRF1 inhibition by STELLA for cancer therapy.
Bai, W., Xu, J., Gu, W., Wang, D., Cui, Y., Rong, W., Du, X., Li, X., Xia, C., Gan, Q., He, G., Guo, H., Deng, J., Wu, Y., Yen, R.C., Yegnasubramanian, S., Rothbart, S.B., Luo, C., Wu, L., Liu, J., Baylin, S.B., Kong, X.(2025) Nat Commun 16: 474-474
- PubMed: 39774694
- DOI: https://doi.org/10.1038/s41467-024-55481-7
- Primary Citation of Related Structures:
8XV4, 8XV6, 8XV7, 8XV8 - PubMed Abstract:
UHRF1 maintains DNA methylation by recruiting DNA methyltransferases to chromatin. In mouse, these dynamics are potently antagonized by a natural UHRF1 inhibitory protein STELLA, while the comparable effects of its human ortholog are insufficiently characterized, especially in cancer cells. Herein, we demonstrate that human STELLA (hSTELLA) is inadequate, while mouse STELLA (mSTELLA) is fully proficient in inhibiting the abnormal DNA methylation and oncogenic functions of UHRF1 in human cancer cells. Structural studies reveal a region of low sequence homology between these STELLA orthologs that allows mSTELLA but not hSTELLA to bind tightly and cooperatively to the essential histone-binding, linked tandem Tudor domain and plant homeodomain (TTD-PHD) of UHRF1, thus mediating ortholog-specific UHRF1 inhibition. For translating these findings to cancer therapy, we use a lipid nanoparticle (LNP)-mediated mRNA delivery approach in which the short mSTELLA, but not hSTELLA regions are required to reverse cancer-specific DNA hypermethylation and impair colorectal cancer tumorigenicity.
Organizational Affiliation:
Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, CAS Key Laboratory of Regenerative Biology, China-New Zealand Joint Laboratory on Biomedicine and Health, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, 510530, China.