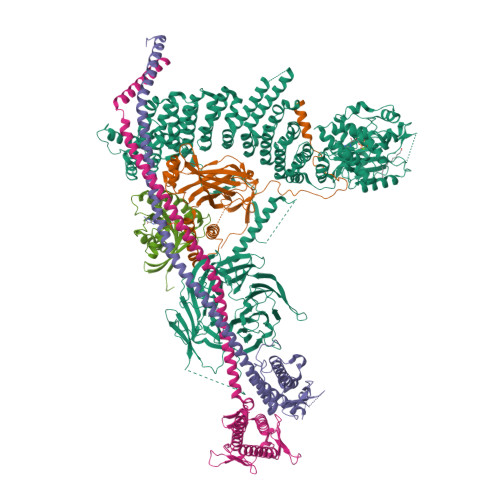
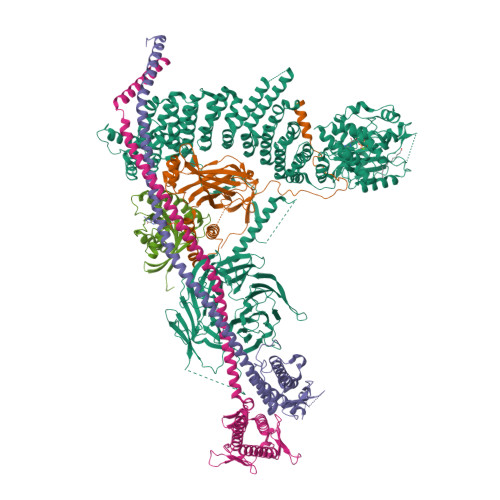
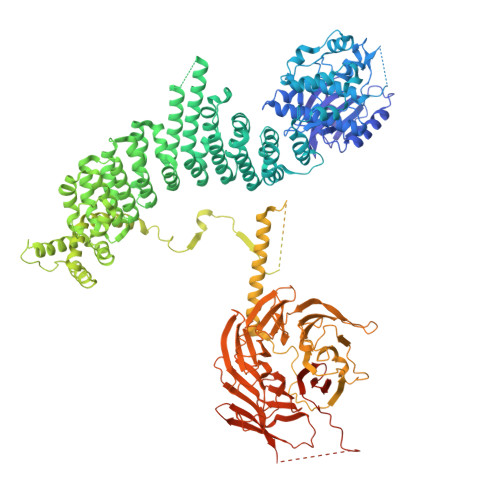
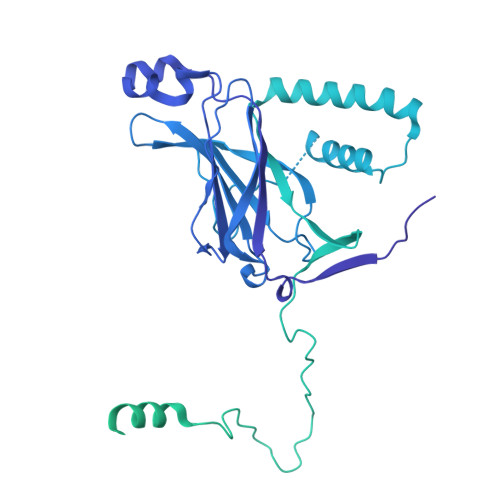
Structural pathway for class III PI 3-kinase activation by the myristoylated GTPbinding pseudokinase VPS15
Cook, A.S.I., Chen, M., Ngyuen, T.N., Claveras-Cabezudo, A., Khuu, G., Rao, S., Garcia, S.N., Yang, M., Iavarone, A.T., Ren, X., Lazarou, M., Hummer, G., Hurley, J.H.To be published.