Full Text |
QUERY: Citation Author = "Law, J.A." | MyPDB Login | Search API |
| Search Summary | This query matches 33 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismTaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateSymmetry TypeSCOP Classification | 1 to 25 of 33 Structures Page 1 of 2 Sort by
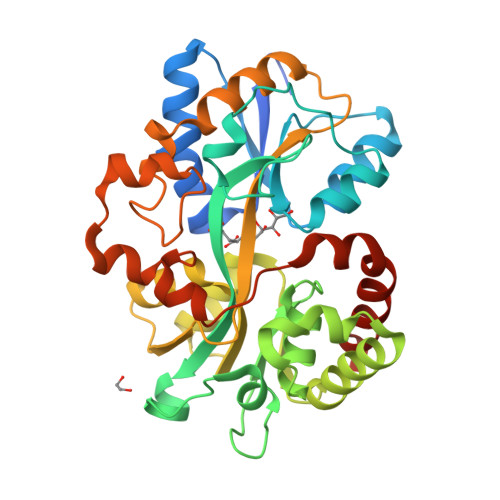
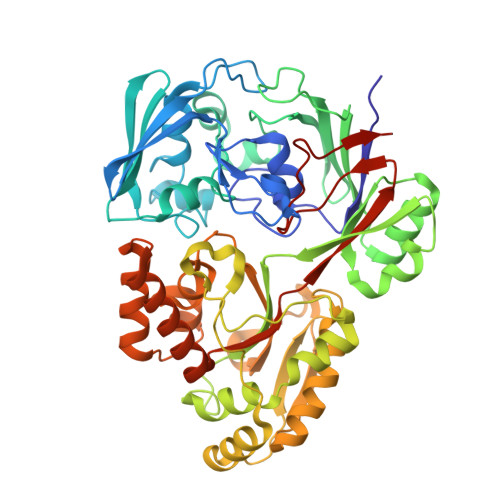
Structure of the PBP MelB (Atu4661) in complex with raffinose from A.fabrum C58(2018) J Biological Chem 293: 7930-7941
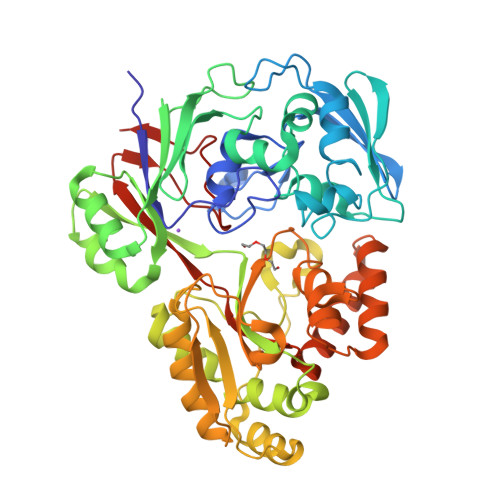
Structure of the periplasmic binding protein MelB (Atu4661) in complex with melibiose from Agrobacterium fabrum C58(2018) J Biological Chem 293: 7930-7941
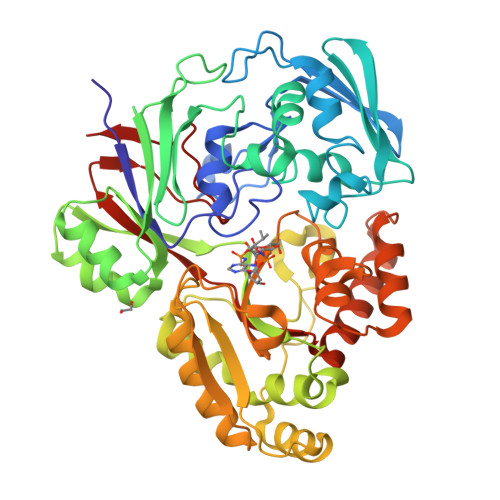
Structure of the periplasmic binding protein (PBP) MelB (atu4661) in complex with galactose from agrobacterium tumefacien C58(2018) J Biological Chem 293: 7930-7941
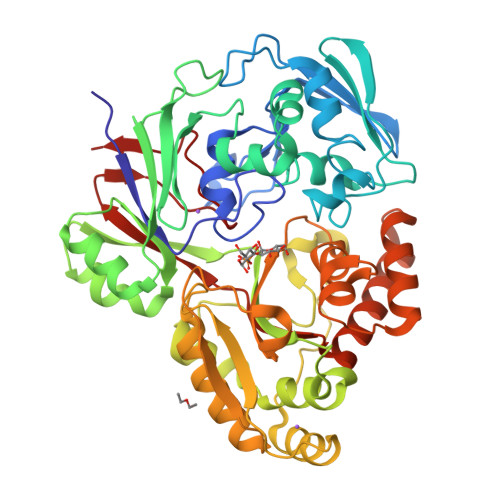
Structure of the periplasmic binding protein (PBP) MelB (Atu4661) in complex with stachyose from agrobacterium fabrum C58(2018) J Biological Chem 293: 7930-7941
Structure of the periplasmic binding protein (PBP) MelB (Atu4661) in complex with galactinol from agrobacterium fabrum C58(2018) J Biological Chem 293: 7930-7941
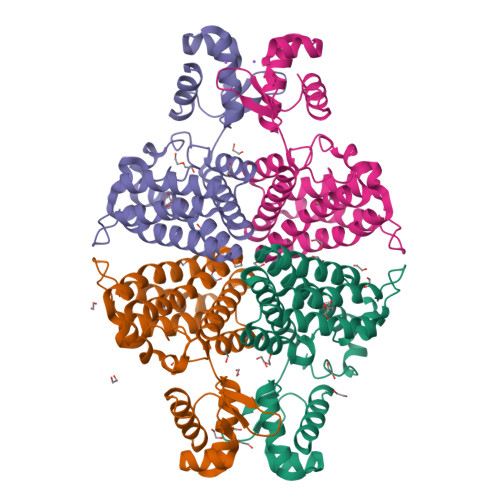
Structure in P3212 form of the PBP/SBP MoaA in complex with mannopinic acid from A.tumefacien R10(2020) Biochem J 477: 615-628
Structure in P3212 form of the PBP/SBP MoaA in complex with glucopinic acid from A.tumefacien R10(2020) Biochem J 477: 615-628
Structure in P21 form of the PBP/SBP MoaA in complex with mannopinic acid from A.tumefacien R10(2020) Biochem J 477: 615-628
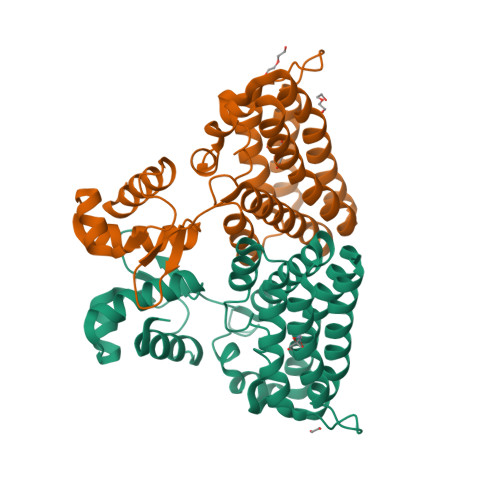
Structure of the PBP/SBP MotA in complex with mannopinic acid from A.tumefacien R10(2020) Biochem J 477: 615-628
Crystal Structure of the PBP/SBP MotA in complex with glucopinic acid from A. tumefaciens B6/R10(2020) Biochem J 477: 615-628
Structure of the transcriptional repressor Atu1419 (VanR) in complex with a fortuitous citrate from agrobacterium fabrum(2021) Nucleic Acids Res 49: 529-546
Structure of the transcriptional repressor Atu1419 (VanR) in complex with a fortuitous citrate from agrobacterium fabrum (P21212 space group)Morera, S., Vigouroux, A., Legrand, P. (2021) Nucleic Acids Res 49: 529-546
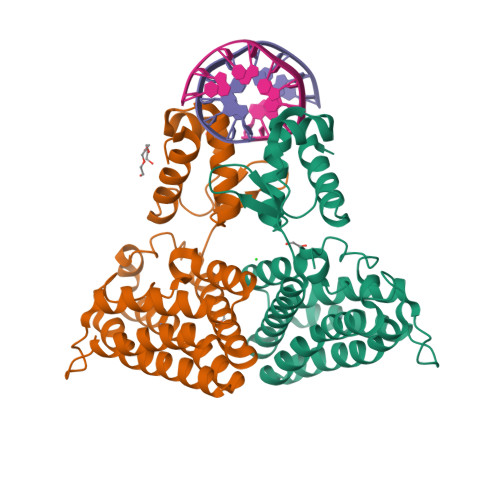
Structure of the transcriptional repressor Atu1419 (VanR) from agrobacterium fabrum in complex a palindromic DNA (C2221 space group)Morera, S., Vigouroux, A., Legrand, P. (2021) Nucleic Acids Res 49: 529-546
Structure of the apo transcriptional repressor Atu1419 (VanR) from agrobacterium fabrumMorera, S., Vigouroux, A., Legrand, P. (2021) Nucleic Acids Res 49: 529-546
Structure of the transcriptional repressor Atu1419 (VanR) from agrobacterium fabrum in complex a palindromic DNA (P6422 space group)Morera, S., Naretto, A., Vigouroux, A., Legrand, P. (2021) Nucleic Acids Res 49: 529-546
PBP AccA from A. tumefaciens Bo542 in apoform 4(2024) Biochem J 481: 93-117
PBP AccA-G145YG440Q mutant from A. tumefaciens Bo542 in apoform 4Morera, S., Vigouroux, A., Legrand, P. (2024) Biochem J 481: 93-117
PBP AccA from A. tumefaciens Bo542 in apoform 1Morera, S., Vigouroux, A., Legrand, P. (2024) Biochem J 481: 93-117
PBP AccA from A. tumefaciens Bo542 in apoform 2Morera, S., Vigouroux, A., legrand, P. (2024) Biochem J 481: 93-117
PBP AccA from A. tumefaciens Bo542 in apoform 3Morera, S., Vigouroux, A., Legrand, P. (2024) Biochem J 481: 93-117
PBP AccA from A. tumefaciens Bo542 in complex with agrocin84Morera, S., Vigouroux, A., El Sahili, A. (2024) Biochem J 481: 93-117
PBP AccA from A. tumefaciens Bo542 in complex with Agrocinopine D-likeMorera, S., Vigouroux, A., Siragu, S. (2024) Biochem J 481: 93-117
PBP AccA from A. tumefaciens Bo542 in complex with D-Glucose-2-phosphate(2024) Biochem J 481: 93-117
PBP AccA-F144YG440Q from A. tumefaciens Bo542 in complex with agrocinopine C-like(2024) Biochem J 481: 93-117
PBP AccA from A. vitis S4 in complex with Agrocinopine A(2024) Biochem J 481: 93-117
1 to 25 of 33 Structures Page 1 of 2 Sort by |