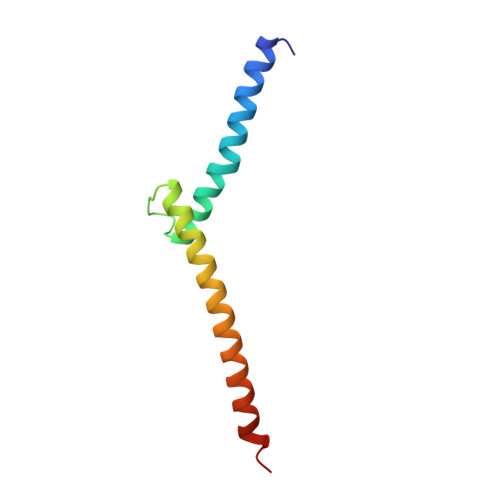
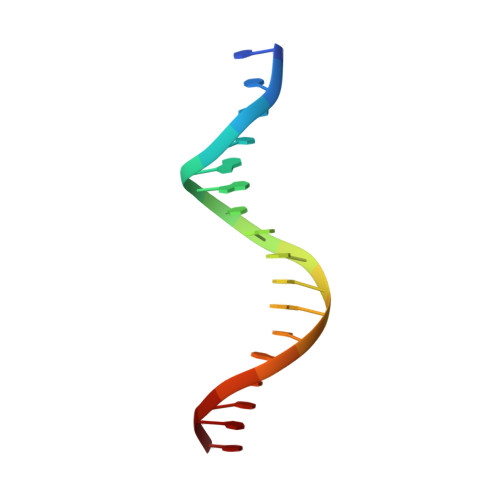
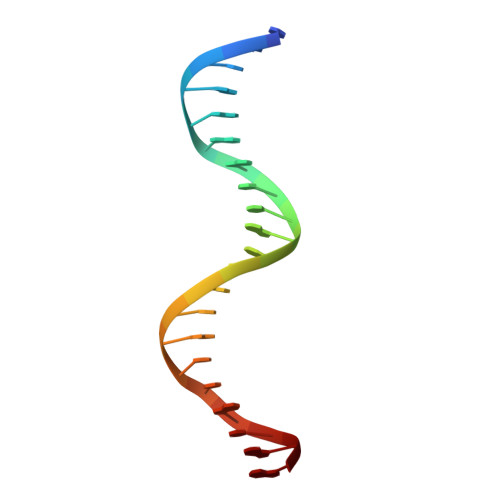
Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 A resolution.
Parraga, A., Bellsolell, L., Ferre-D'Amare, A.R., Burley, S.K.(1998) Structure 6: 661-672
- PubMed: 9634703
- DOI: https://doi.org/10.1016/s0969-2126(98)00067-7
- Primary Citation of Related Structures:
1AM9 - PubMed Abstract:
The sterol regulatory element binding proteins (SREBPs) are helix-loop-helix transcriptional activators that control expression of genes encoding proteins essential for cholesterol biosynthesis/uptake and fatty acid biosynthesis. Unlike helix-loop-helix proteins that recognize symmetric E-boxes (5'-CANNTG-3'), the SREBPs have a tyrosine instead of a conserved arginine in their basic regions. This difference allows recognition of an asymmetric sterol regulatory element (StRE, 5'-ATCACCCAC-3'). The 2.3 A resolution co-crystal structure of the DNA-binding portion of SREBP-1a bound to an StRE reveals a quasi-symmetric homodimer with an asymmetric DNA-protein interface. One monomer binds the E-box half site of the StRE (5'-ATCAC-3') using sidechain-base contacts typical of other helix-loop-helix proteins. The non-E-box half site (5'-GTGGG-3') is recognized through entirely different protein-DNA contacts. Although the SREBPs are structurally similar to the E-box-binding helix-loop-helix proteins, the Arg-->Tyr substitution yields dramatically different DNA-binding properties that explain how they recognize StREs and regulate expression of genes important for membrane biosynthesis.
- Laboratories of Molecular Biophysics, Rockefeller University, New York, NY 10021, USA.
Organizational Affiliation: