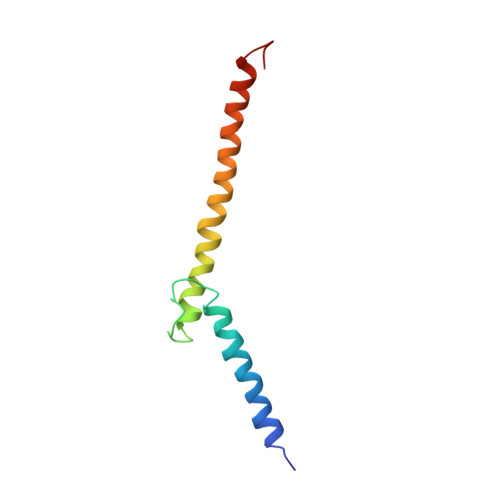
Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain.
Ferre-D'Amare, A.R., Prendergast, G.C., Ziff, E.B., Burley, S.K.(1993) Nature 363: 38-45
- PubMed: 8479534
- DOI: https://doi.org/10.1038/363038a0
- Primary Citation of Related Structures:
1AN2 - PubMed Abstract:
The three-dimensional structure of the basic/helix-loop-helix/leucine zipper domain of the transcription factor Max complexed with DNA has been determined by X-ray crystallography at 2.9 A resolution. Max binds as a dimer to its recognition sequence CACGTG by direct contacts between the alpha-helical basic region and the major groove. This symmetric homodimer, a new protein fold, is a parallel, left-handed, four-helix bundle, with each monomer containing two alpha-helical segments separated by a loop. The two alpha-helical segments are composed of the basic region plus helix 1 and helix 2 plus the leucine repeat, respectively. As in GCN4, the leucine repeat forms a parallel coiled coil.
- Laboratories of Molecular Biophysics, Rockefeller University, New York, New York 10021.
Organizational Affiliation: