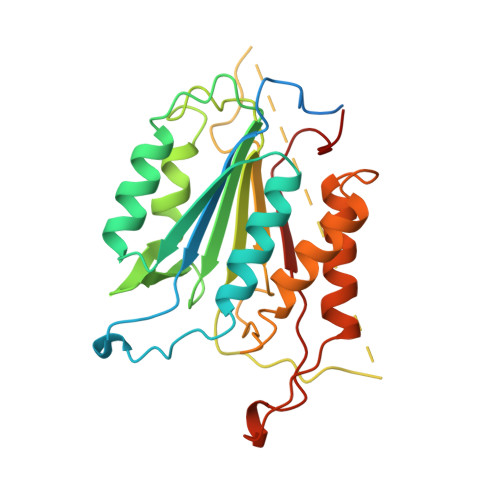
Structure of recombinant human CPP32 in complex with the tetrapeptide acetyl-Asp-Val-Ala-Asp fluoromethyl ketone.
Mittl, P.R., Di Marco, S., Krebs, J.F., Bai, X., Karanewsky, D.S., Priestle, J.P., Tomaselli, K.J., Grutter, M.G.(1997) J Biological Chem 272: 6539-6547
- PubMed: 9045680
- DOI: https://doi.org/10.1074/jbc.272.10.6539
- Primary Citation of Related Structures:
1CP3 - PubMed Abstract:
The cysteine protease CPP32 has been expressed in a soluble form in Escherichia coli and purified to >95% purity. The three-dimensional structure of human CPP32 in complex with the irreversible tetrapeptide inhibitor acetyl-Asp-Val-Ala-Asp fluoromethyl ketone was determined by x-ray crystallography at a resolution of 2.3 A. The asymmetric unit contains a (p17/p12)2 tetramer, in agreement with the tetrameric structure of the protein in solution as determined by dynamic light scattering and size exclusion chromatography. The overall topology of CPP32 is very similar to that of interleukin-1beta-converting enzyme (ICE); however, differences exist at the N terminus of the p17 subunit, where the first helix found in ICE is missing in CPP32. A deletion/insertion pattern is responsible for the striking differences observed in the loops around the active site. In addition, the P1 carbonyl of the ketone inhibitor is pointing into the oxyanion hole and forms a hydrogen bond with the peptidic nitrogen of Gly-122, resulting in a different state compared with the tetrahedral intermediate observed in the structure of ICE and CPP32 in complex with an aldehyde inhibitor. The topology of the interface formed by the two p17/p12 heterodimers of CPP32 is different from that of ICE. This results in different orientations of CPP32 heterodimers compared with ICE heterodimers, which could affect substrate recognition. This structural information will be invaluable for the design of small synthetic inhibitors of CPP32 as well as for the design of CPP32 mutants.
- Core Drug Discovery Technologies, Ciba-Geigy AG, CH-4002 Basel, Switzerland.
Organizational Affiliation: