The crystal structure of human alpha-thrombin complexed with LY178550, a nonpeptidyl, active site-directed inhibitor.
Chirgadze, N.Y., Sall, D.J., Klimkowski, V.J., Clawson, D.K., Briggs, S.L., Hermann, R., Smith, G.F., Gifford-Moore, D.S., Wery, J.P.(1997) Protein Sci 6: 1412-1417
- PubMed: 9232642
- DOI: https://doi.org/10.1002/pro.5560060705
- Primary Citation of Related Structures:
1D4P - PubMed Abstract:
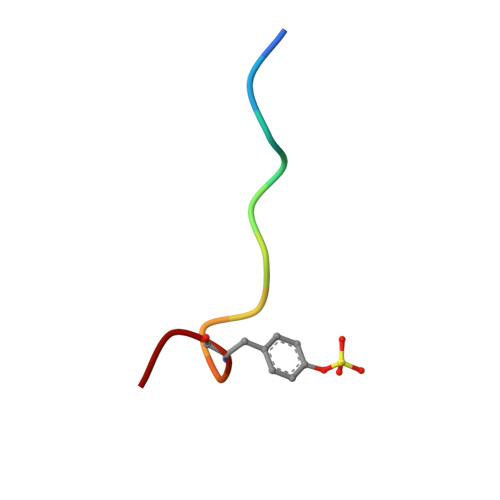
The crystal structure of human alpha-thrombin in complex with LY178550, a nonpeptidyl, active site-directed inhibitor, has been solved to 2.07 A resolution by the method of X-ray crystallography. The final model of the complex has a crystallographic R-value of 21.5% (Rfree = 23.1%) with 0.014 A and 2.4 degrees standard deviation from ideal bond lengths and angles, respectively. Well-defined electron density was observed for the inhibitor in the active site. The inhibitor binds to the active site in an L-shaped manner, mimicking the bound conformation of the tripeptide arginal series of thrombin inhibitors (Chirgadze NY et al., 1992, American Crystallographic Association Meeting 20: 116 [Abstr. PB311]). The basic amidine of LY178550 forms a salt bridge with Asp 189 within the specificity pocket, while the 4-benzylpiperidine side chain engages in a number of hydrophobic interactions at the S2 and S3 binding sites. The inhibitor does not interact in any fashion with the active site sequence Ser 214-Gly 216, as occurs with many of the inhibitors studied previously. The indole N-H of the inhibitor forms a hydrogen bond to the gamma-oxygen of the catalytic serine (Ser 195).
- Lilly Research Laboratories, Indianapolis, Indiana 46285, USA. nyc@lilly.com
Organizational Affiliation: