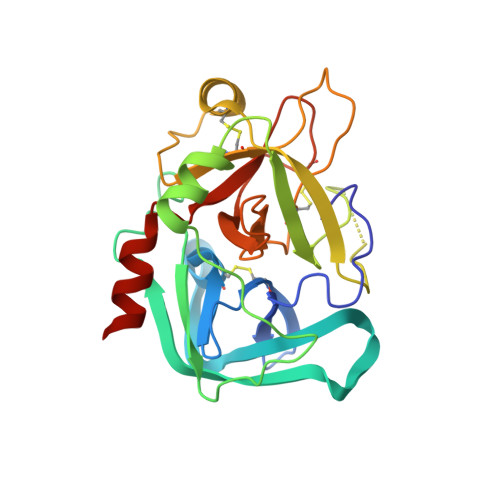
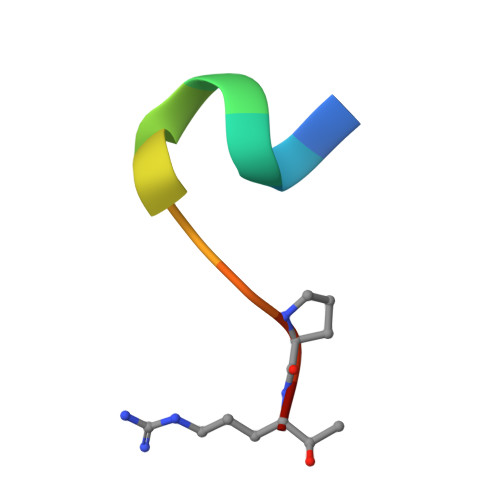
Interaction of the factor XIII activation peptide with alpha -thrombin. Crystal structure of its enzyme-substrate analog complex.
Sadasivan, C., Yee, V.C.(2000) J Biological Chem 275: 36942-36948
- PubMed: 10956659
- DOI: https://doi.org/10.1074/jbc.M006076200
- Primary Citation of Related Structures:
1DE7 - PubMed Abstract:
The serine protease thrombin proteolytically activates blood coagulation factor XIII by cleavage at residue Arg(37); factor XIII in turn cross-links fibrin molecules and gives mechanical stability to the blood clot. The 2.0-A resolution x-ray crystal structure of human alpha-thrombin bound to the factor XIII-(28-37) decapeptide has been determined. This structure reveals the detailed atomic level interactions between the factor XIII activation peptide and thrombin and provides the first high resolution view of this functionally important part of the factor XIII molecule. A comparison of this structure with the crystal structure of fibrinopeptide A complexed with thrombin highlights several important determinants of thrombin substrate interaction. First, the P1 and P2 residues must be compatible with the geometry and chemistry of the S1 and S2 specificity sites in thrombin. Second, a glycine in the P5 position is necessary for the conserved substrate conformation seen in both factor XIII-(28-37) and fibrinopeptide A. Finally, the hydrophobic residues, which occupy the aryl binding site of thrombin determine the substrate conformation further away from the catalytic residues. In the case of factor XIII-(28-37), the aryl binding site is shared by hydrophobic residues P4 (Val(34)) and P9 (Val(29)). A bulkier residue in either of these sites may alter the substrate peptide conformation.
- Department of Molecular Cardiology and Structural Biology Center, Lerner Research Institute, Cleveland Clinic Foundation, Cleveland, Ohio 44195, USA.
Organizational Affiliation: