Structural Basis for the Anticoagulant Activity of the Thrombin-Thrombomodulin Complex
Fuentes-Prior, P., Iwanaga, Y., Huber, R., Pagila, R., Rumennik, G., Seto, M., Morser, J., Light, D.R., Bode, W.(2000) Nature 404: 518
- PubMed: 10761923
- DOI: https://doi.org/10.1038/35006683
- Primary Citation of Related Structures:
1DX5 - PubMed Abstract:
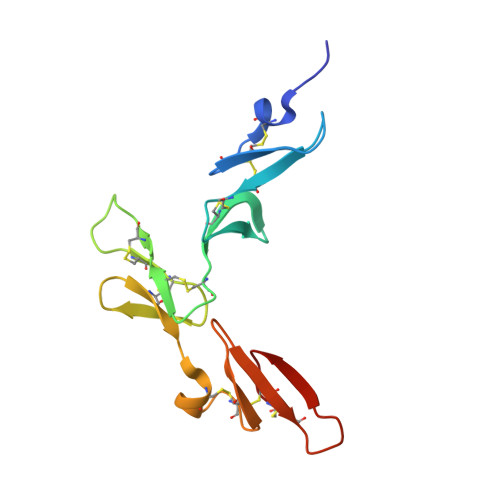
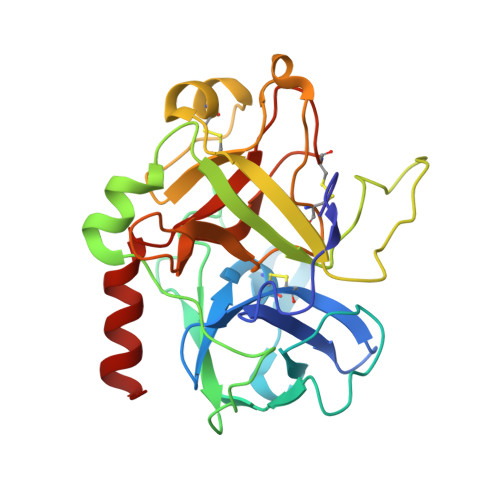
The serine proteinase alpha-thrombin causes blood clotting through proteolytic cleavage of fibrinogen and protease-activated receptors and amplifies its own generation by activating the essential clotting factors V and VIII. Thrombomodulin, a transmembrane thrombin receptor with six contiguous epidermal growth factor-like domains (TME1-6), profoundly alters the substrate specificity of thrombin from pro- to anticoagulant by activating protein C. Activated protein C then deactivates the coagulation cascade by degrading activated factors V and VIII. The thrombin-thrombomodulin complex inhibits fibrinolysis by activating the procarboxypeptidase thrombin-activatable fibrinolysis inhibitor. Here we present the 2.3 A crystal structure of human alpha-thrombin bound to the smallest thrombomodulin fragment required for full protein-C co-factor activity, TME456. The Y-shaped thrombomodulin fragment binds to thrombin's anion-binding exosite-I, preventing binding of procoagulant substrates. Thrombomodulin binding does not seem to induce marked allosteric structural rearrangements at the thrombin active site. Rather, docking of a protein C model to thrombin-TME456 indicates that TME45 may bind substrates in such a manner that their zymogen-activation cleavage sites are presented optimally to the unaltered thrombin active site.
- Max-Planck-Institut für Biochemie, Martinsried, Germany. fuentes@biochem.mpg.de
Organizational Affiliation: