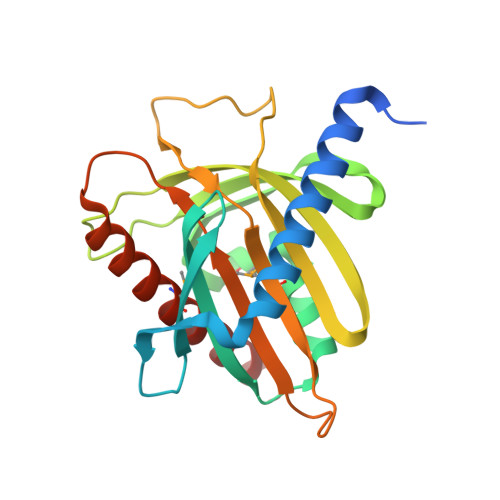
Structure and lipid transport mechanism of a StAR-related domain.
Tsujishita, Y., Hurley, J.H.(2000) Nat Struct Biol 7: 408-414
- PubMed: 10802740
- DOI: https://doi.org/10.1038/75192
- Primary Citation of Related Structures:
1EM2 - PubMed Abstract:
The steroidogenic acute regulatory protein (StAR) regulates acute steroidogenesis in the adrenal cortex and gonads by promoting the translocation of cholesterol to the mitochondrial inner membrane where the first step in steriod biosynthesis is catalyzed. StAR-related lipid transfer (START) domains occur in proteins involved in lipid transport and metabolism, signal transduction, and transcriptional regulation. The 2.2 A resolution crystal structure of the START domain of human MLN64 reported here reveals an alpha/beta fold built around a U-shaped incomplete beta-barrel. The interior of the protein encompasses a 26 x 12 x 11 A hydrophobic tunnel that is large enough to bind a single cholesterol molecule. The StAR and MLN64 START domains bind 1 mole of 14C cholesterol per mole of protein in vitro. Based on the START domain structure and cholesterol binding stoichiometry, it is proposed that StAR acts by shuttling cholesterol molecules one at a time through the intermembrane space of the mitochondrion.
- Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Maryland 20892-0580, USA.
Organizational Affiliation: