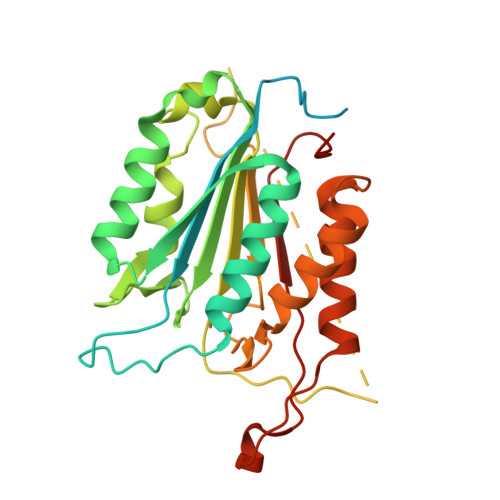
The structures of caspases-1, -3, -7 and -8 reveal the basis for substrate and inhibitor selectivity.
Wei, Y., Fox, T., Chambers, S.P., Sintchak, J., Coll, J.T., Golec, J.M., Swenson, L., Wilson, K.P., Charifson, P.S.(2000) Chem Biol 7: 423-432
- PubMed: 10873833
- DOI: https://doi.org/10.1016/s1074-5521(00)00123-x
- Primary Citation of Related Structures:
1F1J - PubMed Abstract:
Peptide inhibitors of caspases have helped define the role of these cysteine proteases in biology. Structural and biochemical characterization of the caspase enzymes may contribute to the development of new drugs for the treatment of caspase-mediated inflammation and apoptosis. The crystal structure of the previously unpublished caspase-7 (Csp7; 2.35 A) bound to the reversible tetrapeptide aldehyde inhibitor acetyl-Asp-Glu-Val-Asp-CHO is compared with crystal structures of caspases-1 (2.3 A), -3 (2.2 A), and -8 (2.65 A) bound to the same inhibitor. Csp7 is a close homolog of caspase-3 (Csp3), and these two caspases possess some quarternary structural characteristics that support their unique role among the caspase family. However, although Csp3 and Csp7 are quite similar overall, they were found to have a significantly different substitution pattern of amino acids in and around the S4-binding site. These structures span all three caspase subgroups, and provide a basis for inferring substrate and inhibitor binding, as well as selectivity for the entire caspase family. This information will influence the design of selective caspase inhibitors to further elucidate the role of caspases in biology and hopefully lead to the design of therapeutic agents to treat caspase-mediated diseases, such as rheumatoid arthritis, certain neurogenerative diseases and stroke.
- Vertex Pharmaceuticals, Cambridge, MA 02139-4242, USA.
Organizational Affiliation: