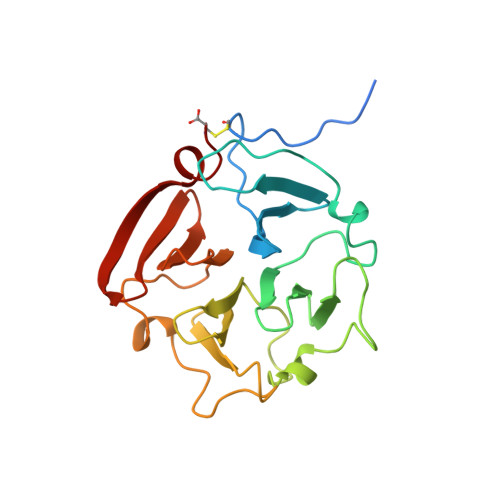
Crystal structure of the haemopexin-like C-terminal domain of gelatinase A.
Libson, A.M., Gittis, A.G., Collier, I.E., Marmer, B.L., Goldberg, G.I., Lattman, E.E.(1995) Nat Struct Biol 2: 938-942
- PubMed: 7583664
- DOI: https://doi.org/10.1038/nsb1195-938
- Primary Citation of Related Structures:
1GEN - PubMed Abstract:
The crystal structure of the haemopexin-like C-terminal domain of gelatinase A reveals that it is a four-bladed beta-propeller protein. The four blades are arranged around a channel-like opening in which Ca2+ and a Na-Cl+ ion pair are bound.