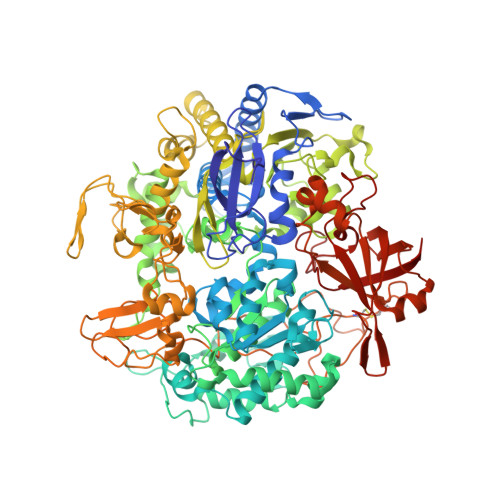
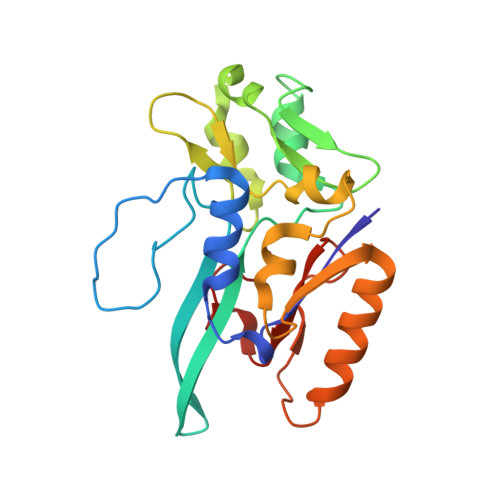
Gene Sequence and the 1.8 A Crystal Structure of the Tungsten-Containing Formate Dehydrogenase from Desulfovibrio Gigas
Raaijmakers, H.C.A., Macieira, S., Dias, J., Teixeira, S., Bursakov, S., Huber, R., Moura, J., Moura, I., Romao, M.(2002) Structure 10: 1261
- PubMed: 12220497
- DOI: https://doi.org/10.1016/s0969-2126(02)00826-2
- Primary Citation of Related Structures:
1H0H - PubMed Abstract:
Desulfovibrio gigas formate dehydrogenase is the first representative of a tungsten-containing enzyme from a mesophile that has been structurally characterized. It is a heterodimer of 110 and 24 kDa subunits. The large subunit, homologous to E. coli FDH-H and to D. desulfuricans nitrate reductase, harbors the W site and one [4Fe-4S] center. No small subunit ortholog containing three [4Fe-4S] clusters has been reported. The structural homology with E. coli FDH-H shows that the essential residues (SeCys158, His159, and Arg407) at the active site are conserved. The active site is accessible via a positively charged tunnel, while product release may be facilitated, for H(+) by buried waters and protonable amino acids and for CO(2) through a hydrophobic channel.
- REQUIMTE/CQFB, Departamento de Química, FCT, Universidade Nova de Lisboa, 2829-516 Caparica, Portugal.
Organizational Affiliation: