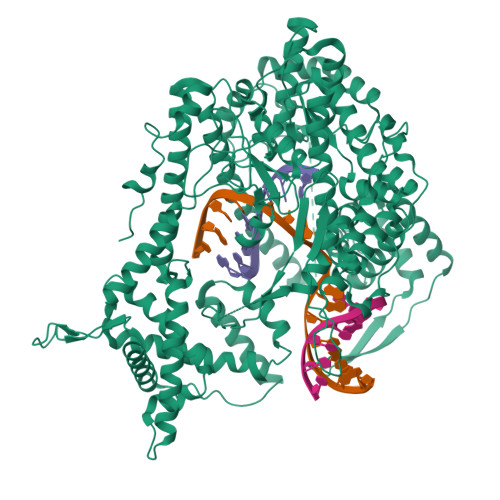
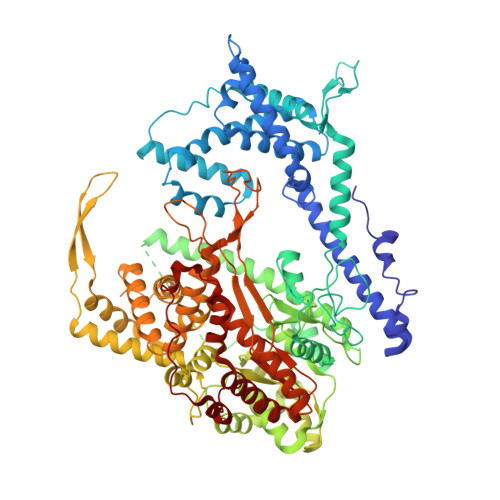
Structure of a T7 RNA Polymerase Elongation Complex at 2.9 A Resolution
Tahirov, T.H., Temiakov, D., Anikin, M., Patlan, V., Mcallister, W.T., Vassylyev, D.G., Yokoyama, S.(2002) Nature 420: 43
- PubMed: 12422209
- DOI: https://doi.org/10.1038/nature01129
- Primary Citation of Related Structures:
1H38 - PubMed Abstract:
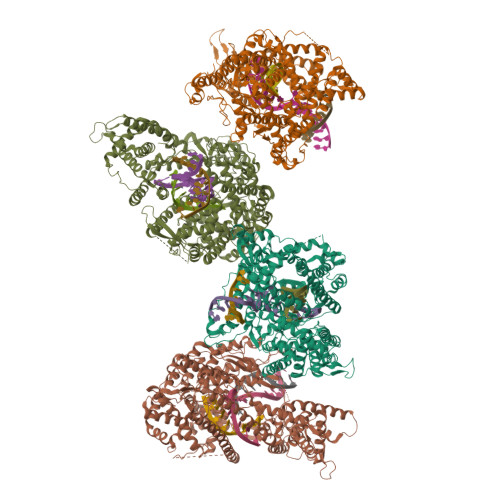
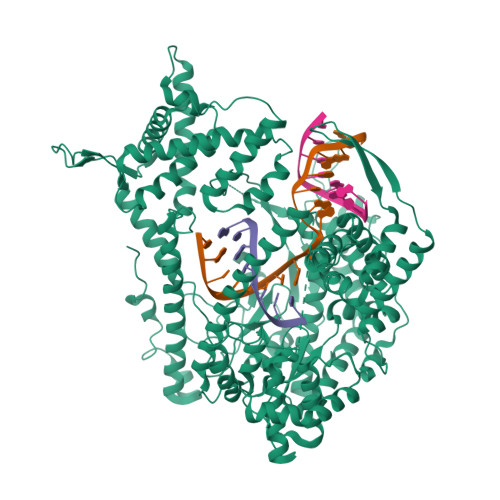
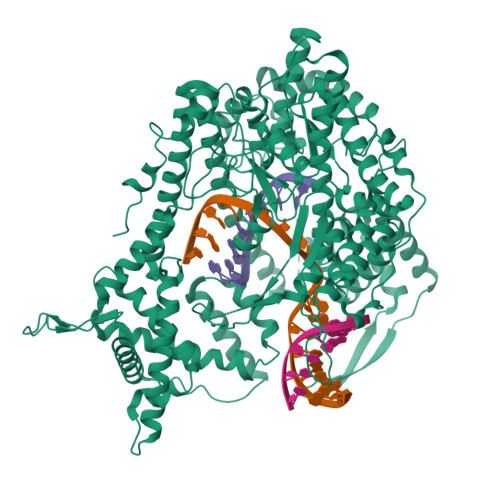
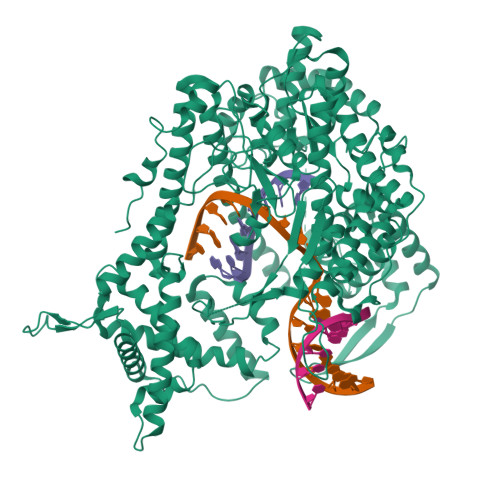
The single-subunit bacteriophage T7 RNA polymerase carries out the transcription cycle in an identical manner to that of bacterial and eukaryotic multisubunit enzymes. Here we report the crystal structure of a T7 RNA polymerase elongation complex, which shows that incorporation of an 8-base-pair RNA-DNA hybrid into the active site of the enzyme induces a marked rearrangement of the amino-terminal domain. This rearrangement involves alternative folding of about 130 residues and a marked reorientation (about 130 degrees rotation) of a stable core subdomain, resulting in a structure that provides elements required for stable transcription elongation. A wide opening on the enzyme surface that is probably an RNA exit pathway is formed, and the RNA-DNA hybrid is completely buried in a newly formed, deep protein cavity. Binding of 10 base pairs of downstream DNA is stabilized mostly by long-distance electrostatic interactions. The structure implies plausible mechanisms for the various phases of the transcription cycle, and reveals important structural similarities with the multisubunit RNA polymerases.
Organizational Affiliation:
High Throughput Factory, RIKEN Harima Institute at SPring-8, 1-1-1 Kouto, Mikazuki-cho, Sayo, Hyogo 679-5148, Japan.