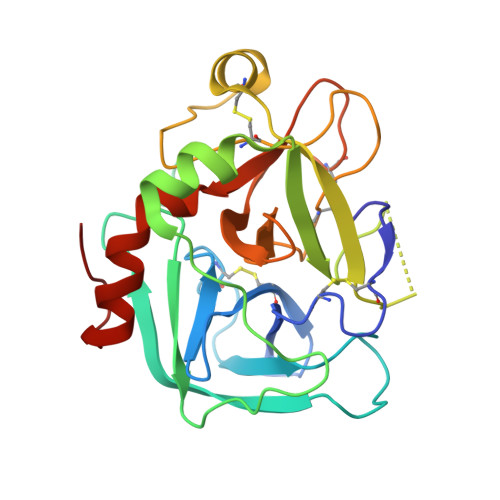
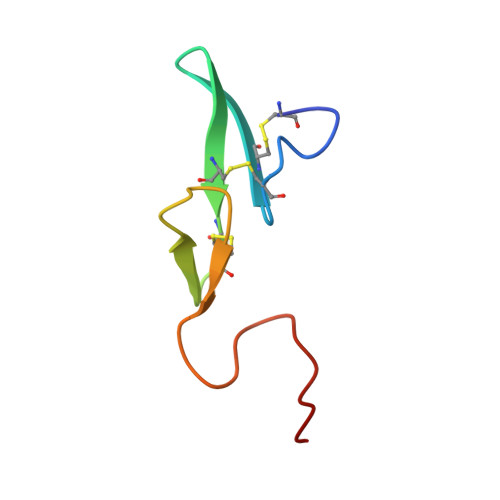
Structure of human des(1-45) factor Xa at 2.2 A resolution.
Padmanabhan, K., Padmanabhan, K.P., Tulinsky, A., Park, C.H., Bode, W., Huber, R., Blankenship, D.T., Cardin, A.D., Kisiel, W.(1993) J Mol Biology 232: 947-966
- PubMed: 8355279
- DOI: https://doi.org/10.1006/jmbi.1993.1441
- Primary Citation of Related Structures:
1HCG - PubMed Abstract:
The structure of a large molecular fragment of factor Xa that lacks only a Gla (gamma-carboxyglutamic acid) domain (N-terminal 45 residues) has been solved by X-ray crystallography and refined at 2.2 A resolution to a crystallographic R-value of 0.168. The fragment identity was clearly established by automated Edman degradation. X-ray structure analysis confirmed the biochemical characterization and also revealed that the N-terminal epidermal growth factor (EGF)-like domain is flexibly disordered in crystals. The second EGF module, however, is positionally ordered making contacts with the catalytic domain. The overall folding of the catalytic domain is similar to that of alpha-thrombin, excluding the insertion loops of the latter with respect to simpler serine proteinases. The C-terminal arginine of the A-chain interacts in a substrate-like manner with the S1 specificity site of the active site of a crystallographically neighboring molecule. Based on this interaction and the structure of D-PheProArg methylene-thrombin, a model of the commonly used dansylGluGlyArg methylene inhibitor-factor Xa interaction is proposed. The region of factor Xa corresponding to the fibrinogen recognition site of thrombin has a reversed electrical polarity to the anion binding fibrinogen recognition site of thrombin but possesses a site similar to the Ca2+ binding site of trypsin and other serine proteinases. The structure of the C-terminal EGF domain of factor Xa is the first to be determined crystallographically. Its folding has been comprehensively compared with similar domains determined by NMR. Although the A-chain makes 44 contacts at less than 3.5 A with the catalytic domain, only 16 involve the EGF module. In addition, the A-chain makes 30 intermolecular contacts with a neighboring catalytic domain.
- Department of Chemistry, Michigan State University, East Lansing 48824-1322.
Organizational Affiliation: