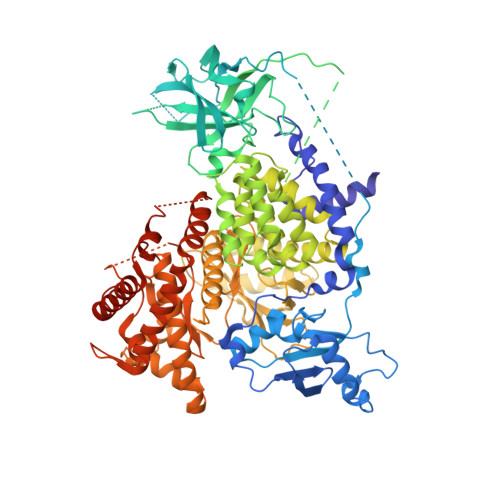
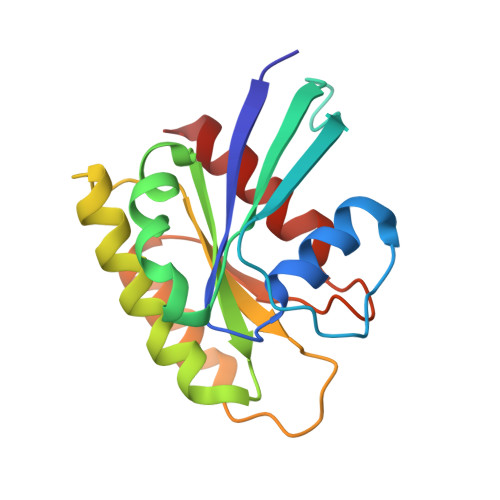
Crystal Structure and Functional Analysis of Ras Binding to its Effector Phosphoinositide 3-Kinase Gamma
Pacold, M.E., Suire, S., Perisic, O., Lara-Gonzalez, S., Davis, C.T., Walker, E.H., Hawkins, P.T., Stephens, L., Eccleston, J.F., Williams, R.L.(2000) Cell 103: 931
- PubMed: 11136978
- DOI: https://doi.org/10.1016/s0092-8674(00)00196-3
- Primary Citation of Related Structures:
1HE8 - PubMed Abstract:
Ras activation of phosphoinositide 3-kinase (PI3K) is important for survival of transformed cells. We find that PI3Kgamma is strongly and directly activated by H-Ras G12V in vivo or by GTPgammaS-loaded H-Ras in vitro. We have determined a crystal structure of a PI3Kgamma/Ras.GMPPNP complex. A critical loop in the Ras binding domain positions Ras so that it uses its switch I and switch II regions to bind PI3Kgamma. Mutagenesis shows that interactions with both regions are essential for binding PI3Kgamma. Ras also forms a direct contact with the PI3Kgamma catalytic domain. These unique Ras/PI3Kgamma interactions are likely to be shared by PI3Kalpha. The complex with Ras shows a change in the PI3K conformation that may represent an allosteric component of Ras activation.
- MRC Laboratory of Molecular Biology Hills Road CB2 2QH, Cambridge, United Kingdom.
Organizational Affiliation: