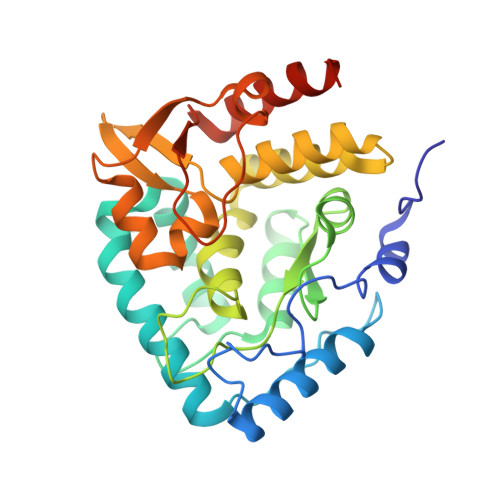
Three-dimensional structure of human tryptophan hydroxylase and its implications for the biosynthesis of the neurotransmitters serotonin and melatonin
Wang, L., Erlandsen, H., Haavik, J., Knappskog, P.M., Stevens, R.C.(2002) Biochemistry 41: 12569-12574
- PubMed: 12379098
- DOI: https://doi.org/10.1021/bi026561f
- Primary Citation of Related Structures:
1MLW - PubMed Abstract:
Tryptophan hydroxylase oxidizes L-tryptophan to 5-hydroxy-L-tryptophan in the rate-determining step of serotonin biosynthesis. We have determined the X-ray crystal structure (1.7 A) of a truncated functional form of human tryptophan hydroxylase with the bound cofactor analogue 7,8-dihydro-L-biopterin, providing the first atomic-resolution information for the catalytic domain of this important enzyme. Comparison of the three-dimensional structures of all three members of the aromatic amino acid hydroxylase family--tyrosine hydroxylase, phenylalanine hydroxylase, and tryptophan hydroxylase--reveals important differences at the active sites.
- Department of Molecular Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, California 92037, USA.
Organizational Affiliation: