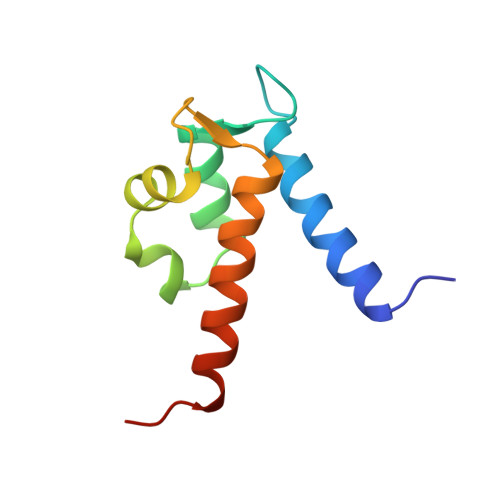
The structure of human MRP8, a member of the S100 calcium-binding protein family, by MAD phasing at 1.9 A resolution.
Ishikawa, K., Nakagawa, A., Tanaka, I., Suzuki, M., Nishihira, J.(2000) Acta Crystallogr D Biol Crystallogr 56: 559-566
- PubMed: 10771424
- DOI: https://doi.org/10.1107/s0907444900002833
- Primary Citation of Related Structures:
1MR8 - PubMed Abstract:
The structure of human MRP8 in the calcium-bound form was determined at 1.9 A resolution by X-ray crystallography. The structure was initially solved by MAD phasing of an ytterbium-substituted crystal and was refined against data obtained from a Ca(2+)-bound crystal. The dimeric form of MRP8 was stabilized by hydrophobic interactions between mutually wrapped helices. There were two EF-hand motifs per monomer and each EF-hand bound one Ca(2+) with a different affinity [the affinity of the C-terminal EF-hand (EF-2) for Ca(2+) was stronger than that of the N-terminal EF-hand (EF-1)]. Furthermore, replacement with Yb(3+) occurred in the C-terminal EF-hand only, suggesting a more flexible nature for EF-2 than for EF-1. This, combined with previous observations that the helix in EF-2 (helix III) undergoes a large conformational change upon calcium binding, suggests that the C-terminal EF-hand (EF-2) plays a role as a trigger for Ca(2+)-induced conformational change.
- Division of Biological Sciences, Graduate School of Science, Hokkaido University, Sapporo 060-0810, Japan.
Organizational Affiliation: