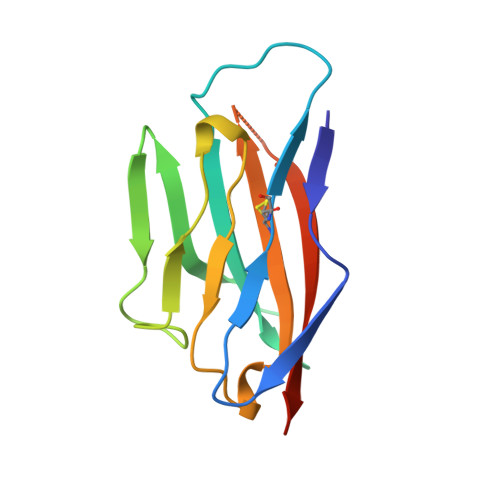
Crystal structure of the extracellular domain from P0, the major structural protein of peripheral nerve myelin.
Shapiro, L., Doyle, J.P., Hensley, P., Colman, D.R., Hendrickson, W.A.(1996) Neuron 17: 435-449
- PubMed: 8816707
- DOI: https://doi.org/10.1016/s0896-6273(00)80176-2
- Primary Citation of Related Structures:
1NEU - PubMed Abstract:
P0, the major protein of peripheral nerve myelin, mediates membrane adhesion in the spiral wraps of the myelin sheath. We have determined the crystal structure of the extracellular domain from P0 (P0ex) at 1.9 A resolution. P0ex is folded like a typical immunoglobulin variable-like domain; five residues at the C-terminus are disordered, suggesting a flexible linkage to the membrane. The requirements for crystallization of P0ex are similar to those for maintaining the native extracellular spacing of adjacent myelin lamellae; thus, given the self-adhesive character of P0ex, the crystal itself may reveal some of the natural interactions that occur between P0 molecules in myelin. The structure leads to the suggestion that P0 extracellular domains may emanate from the membrane surface as tetramers that link to tetramers on the opposing membrane surface, to result in the formation of networks of molecules. We report analytical ultracentrifugation data for P0ex that support this idea.
- Department of Biochemistry and Molecular Biophysics, Columbia University, New York, New York 10032, USA.
Organizational Affiliation: