Molecular Recognition of Paxillin LD Motifs by the Focal Adhesion Targeting Domain
Hoellerer, M.K., Noble, M.E.M., Labesse, G., Werner, J.M., Arold, S.T.(2003) Structure 11: 1207-1217
- PubMed: 14527389
- DOI: https://doi.org/10.1016/j.str.2003.08.010
- Primary Citation of Related Structures:
1OW6, 1OW7, 1OW8 - PubMed Abstract:
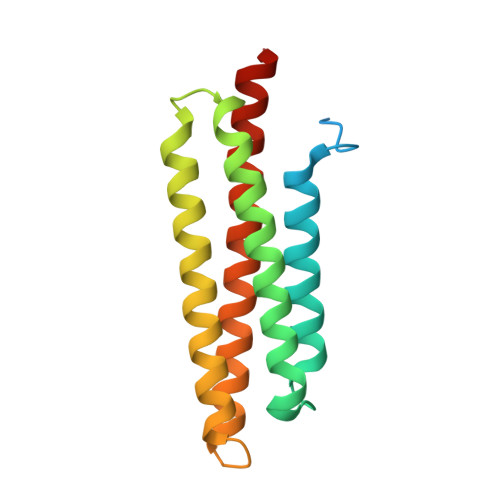
Focal adhesions (FAs) are large submembrane signaling complexes formed at sites of cellular attachment to the extracellular matrix. The interaction of LD motifs with their targets plays an important role in the assembly of FAs. We have determined the molecular basis for the recognition of two paxillin LD motifs by the FA targeting (FAT) domain of FA kinase using a combination of X-ray crystallography, solution NMR, and homology modeling. The four-helix FAT domain displays two LD binding sites on opposite sites of the molecule that bind LD peptides in a helical conformation. Threading studies suggest that the LD-interacting domain of p95PKL shares a common four-helical core with the FAT domain and the tail of vinculin, defining a structural family of LD motif binding modules.
- Biochemistry Department, South Parks Road, Oxford, OX1 3QU, United Kingdom.
Organizational Affiliation: