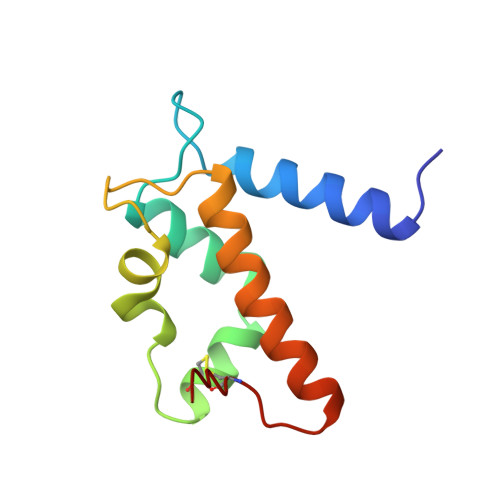
EF-hands at atomic resolution: the structure of human psoriasin (S100A7) solved by MAD phasing.
Brodersen, D.E., Etzerodt, M., Madsen, P., Celis, J.E., Thogersen, H.C., Nyborg, J., Kjeldgaard, M.(1998) Structure 6: 477-489
- PubMed: 9562557
- DOI: https://doi.org/10.1016/s0969-2126(98)00049-5
- Primary Citation of Related Structures:
1PSR - PubMed Abstract:
The S100 family consists of small acidic proteins, belonging to the EF-hand class of calcium-binding proteins. They are primarily regulatory proteins, involved in cell growth, cell structure regulation and signal transduction. Psoriasin (S100A7) is an 11.7 kDa protein that is highly upregulated in the epidermis of patients suffering from the chronic skin disease psoriasis. Although its exact function is not known, psoriasin is believed to participate in the biochemical response which follows transient changes in the cellular Ca2+ concentration. The three-dimensional structure of holmium-substituted psoriasin has been determined by multiple anomalous wavelength dispersion (MAD) phasing and refined to atomic resolution (1.05 A). The structure represents the most accurately determined structure of a calcium-binding protein. Although the overall structure of psoriasin is similar to those of other S100 proteins, several important differences exist, mainly in the N-terminal EF-hand motif that contains a distorted loop and lacks a crucial calcium-binding residue. It is these minor differences that may account for the different specificities among members of this family. The structure of human psoriasin reveals that this protein, in contrast to other S100 proteins with known structure, is not likely to strongly bind more than one calcium ion per monomer. The present study contradicts the idea that calcium binding induces large changes in conformation, as suggested by previously determined structures of apo forms of S100 proteins. The substitution of Ca2+ ions in EF-hands by lanthanide ions may provide a general vehicle for structure determination of S100 proteins by means of MAD phasing.
- Macromolecular Crystallography, Aarhus University, Gustav Wieds Vej 10c, DK-8000, Aarhus C, Denmark.
Organizational Affiliation: