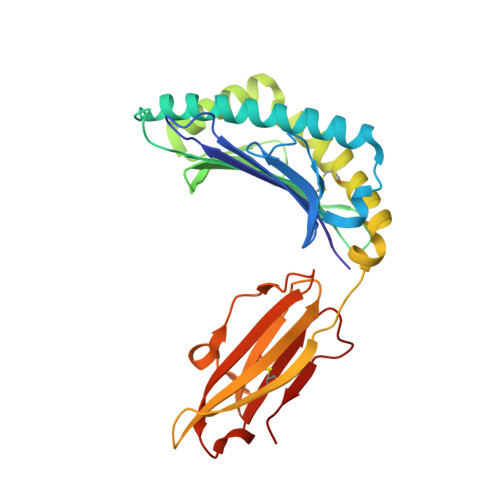
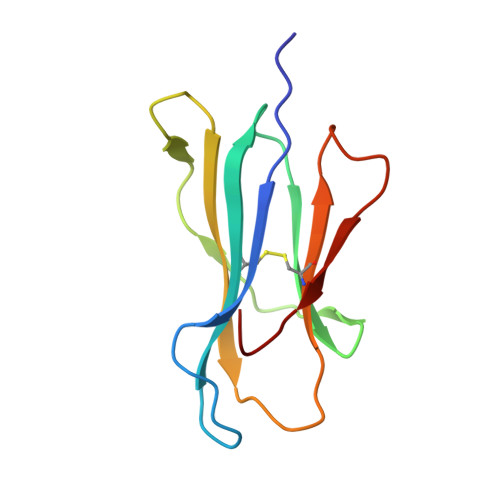
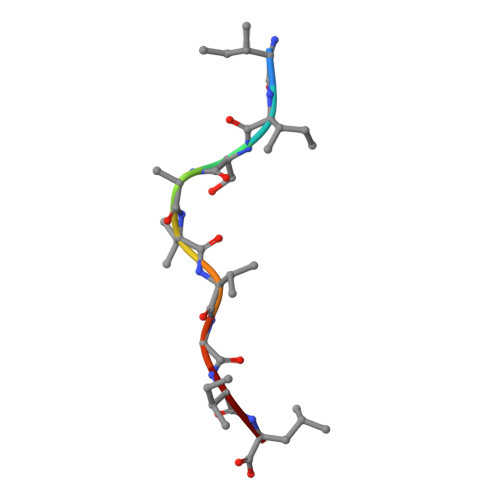
Poor binding of a HER-2/neu epitope (GP2) to HLA-A2.1 is due to a lack of interactions with the center of the peptide.
Kuhns, J.J., Batalia, M.A., Yan, S., Collins, E.J.(1999) J Biological Chem 274: 36422-36427
- PubMed: 10593938
- DOI: https://doi.org/10.1074/jbc.274.51.36422
- Primary Citation of Related Structures:
1QR1 - PubMed Abstract:
Class I major histocompatibility complex (MHC) molecules bind short peptides derived from proteins synthesized within the cell. These complexes of peptide and class I MHC (pMHC) are transported from the endoplasmic reticulum to the cell surface. If a clonotypic T cell receptor expressed on a circulating T cell binds to the pMHC complex, the cell presenting the pMHC is killed. In this manner, some tumor cells expressing aberrant proteins are recognized and removed by the immune system. However, not all tumors are recognized efficiently. One reason hypothesized for poor T cell recognition of tumor-associated peptides is poor binding of those peptides to class I MHC molecules. Many peptides, derived from the proto-oncogene HER-2/neu have been shown to be recognized by cytotoxic T cells derived from HLA-A2(+) patients with breast cancer and other adenocarcinomas. Seven of these peptides were found to bind with intermediate to poor affinity. In particular, GP2 (HER-2/neu residues 654-662) binds very poorly even though it is predicted to bind well based upon the presence of the correct HLA-A2.1 peptide-binding motif. Altering the anchor residues to those most favored by HLA-A2.1 did not significantly improve binding affinity. The crystallographic structure shows that unlike other class I-peptide structures, the center of the peptide does not assume one specific conformation and does not make stabilizing contacts with the peptide-binding cleft.
- Department of Microbiology and Immunology, University of North Carolina, Chapel Hill, North Carolina 27599, USA.
Organizational Affiliation: