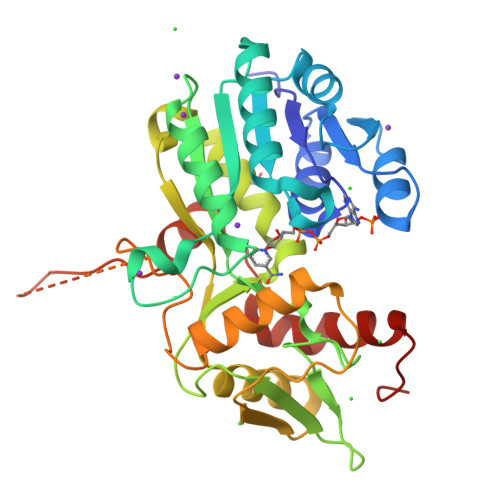
The negative transcriptional regulator NmrA discriminates between oxidized and reduced dinucleotides.
Lamb, H.K., Leslie, K., Dodds, A.L., Nutley, M., Cooper, A., Johnson, C., Thompson, P., Stammers, D.K., Hawkins, A.R.(2003) J Biol Chem 278: 32107-32114
- PubMed: 12764138
- DOI: https://doi.org/10.1074/jbc.M304104200
- Primary Citation of Related Structures:
1TI7 - PubMed Abstract:
NmrA, a transcription repressor involved in the regulation of nitrogen metabolism in Aspergillus nidulans,is a member of the short-chain dehydrogenase reductase superfamily. Isothermal titration calorimetry and differential scanning calorimetry have been used to show NmrA binds NAD+ and NADP+ with similar affinity (average KD 65 microM) but has a greatly reduced affinity for NADH and NADPH (average KD 6.0 mM). The structure of NmrA in a complex with NADP+ reveals how repositioning a His-37 side chain allows the different conformations of NAD+ and NADP+ to be accommodated. Modeling NAD(P)H into NmrA indicated that steric clashes, attenuation of electrostatic interactions, and loss of aromatic ring stacking can explain the differing affinities of NAD(P)+/NAD(P)H. The ability of NmrA to discriminate between the oxidized and reduced forms of the dinucleotides may be linked to a possible role in redox sensing. Isothermal titration calorimetry demonstrated that NmrA and a C-terminal fragment of the GATA transcription factor AreA interacted with a 1:1 stoichiometry and an apparent KD of 0.26 microM. NmrA was unable to bind the nitrogen metabolite repression signaling molecules ammonium or glutamine.
Organizational Affiliation:
School of Cell and Molecular Biosciences, Catherine Cookson Building, University of Newcastle upon Tyne, Framlington Place, NE2 4HH, United Kingdom.