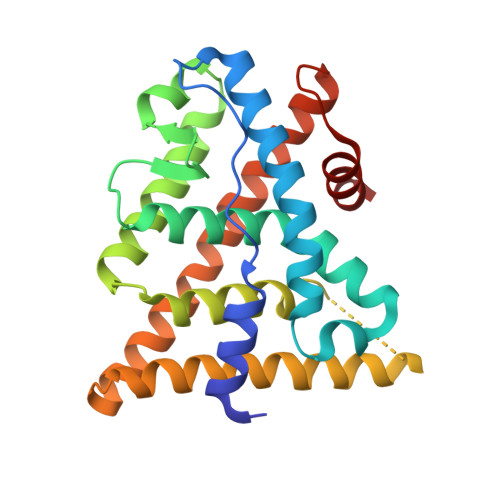
Design and synthesis of aryl diphenolic azoles as potent and selective estrogen receptor-beta ligands.
Malamas, M.S., Manas, E.S., McDevitt, R.E., Gunawan, I., Xu, Z.B., Collini, M.D., Miller, C.P., Dinh, T., Henderson, R.A., Keith Jr., J.C., Harris, H.A.(2004) J Med Chem 47: 5021-5040
- PubMed: 15456246
- DOI: https://doi.org/10.1021/jm049719y
- Primary Citation of Related Structures:
1U3Q, 1U3R, 1U3S - PubMed Abstract:
New diphenolic azoles as highly selective estrogen receptor-beta agonists are reported. The more potent and selective analogues of these series have comparable binding affinities for ERbeta as the natural ligand 17beta-estradiol but are >100-fold selective over ERalpha. Our design strategy not only followed a traditional SAR approach but also was supported by X-ray structures of ERbeta cocrystallized with various ligands as well as molecular modeling studies. These strategies enabled us to take advantage of a single conservative residue substitution in the ligand-binding pocket, ERalpha Met(421) --> ERbeta Ile(373), to optimize ERbeta selectivity. The 7-position-substituted benzoxazoles (Table 5) were the most selective ligands of both azole series, with ERB-041 (117) being >200-fold selective for ERbeta. The majority of ERbeta selective agonists tested that were at least approximately 50-fold selective displayed a consistent in vivo profile: they were inactive in several models of classic estrogen action (uterotrophic, osteopenia, and vasomotor instability models) and yet were active in the HLA-B27 transgenic rat model of inflammatory bowel disease. These data suggest that ERbeta-selective agonists are devoid of classic estrogenic effects and may offer a novel therapy to treat certain inflammatory conditions.
- Department of Chemical and Screening Sciences, Wyeth Research, CN 8000, Princeton, NJ 08543-8000, USA. malamam@wyeth.com
Organizational Affiliation: