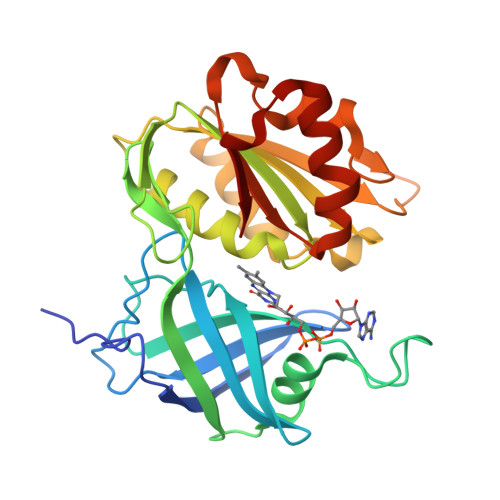
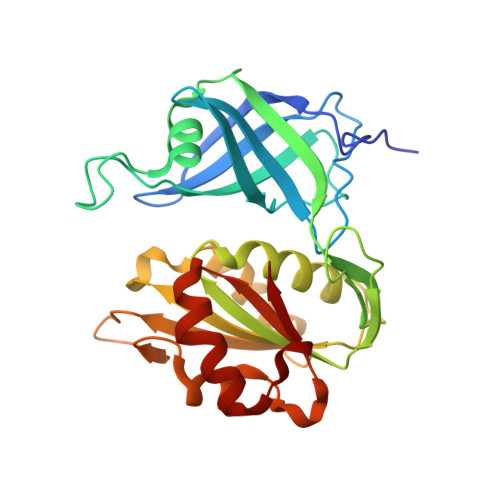
Structure of human erythrocyte NADH-cytochrome b5 reductase.
Bando, S., Takano, T., Yubisui, T., Shirabe, K., Takeshita, M., Nakagawa, A.(2004) Acta Crystallogr D Biol Crystallogr 60: 1929-1934
- PubMed: 15502298
- DOI: https://doi.org/10.1107/S0907444904020645
- Primary Citation of Related Structures:
1UMK - PubMed Abstract:
Erythrocyte NADH-cytochrome b(5) reductase reduces methaemoglobin to functional haemoglobin. In order to examine the function of the enzyme, the structure of NADH-cytochrome b(5) reductase from human erythrocytes has been determined and refined by X-ray crystallography. At 1.75 A resolution, the root-mean-square deviations (r.m.s.d.) from standard bond lengths and angles are 0.006 A and 1.03 degrees , respectively. The molecular structure was compared with those of rat NADH-cytochrome b(5) reductase and corn nitrate reductase. The human reductase resembles the rat reductase in overall structure as well as in many side chains. Nevertheless, there is a large main-chain shift from the human reductase to the rat reductase or the corn reductase caused by a single-residue replacement from proline to threonine. A model of the complex between cytochrome b(5) and the human reductase has been built and compared with that of the haem-containing domain of the nitrate reductase molecule. The interaction between cytochrome b(5) and the human reductase differs from that of the nitrate reductase because of differences in the amino-acid sequences. The structures around 15 mutation sites of the human reductase have been examined for the influence of residue substitutions using the program ROTAMER. Five mutations in the FAD-binding domain seem to be related to cytochrome b(5).
Organizational Affiliation:
Institute for Protein Research, Osaka University, 3-2 Yamadaoka, Suita, Osaka 565-0871, Japan.