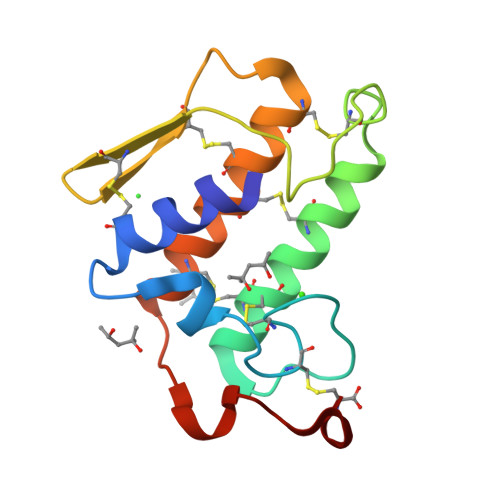
A redetermination of the structure of the triple mutant (K53,56,120M) of phospholipase A2 at 1.6 A resolution using sulfur-SAS at 1.54 A wavelength.
Sekar, K., Rajakannan, V., Velmurugan, D., Yamane, T., Thirumurugan, R., Dauter, M., Dauter, Z.(2004) Acta Crystallogr D Biol Crystallogr 60: 1586-1590
- PubMed: 15333929
- DOI: https://doi.org/10.1107/S090744490401697X
- Primary Citation of Related Structures:
1VKQ - PubMed Abstract:
The crystal structure of the triple mutant K53,56,120M of bovine pancreatic phospholipase A(2) has been redetermined using sulfur single-wavelength anomalous scattering. The synchrotron data were collected at lambda = 1.54 A and the crystal diffracted to 1.6 A resolution. The program SOLVE was used to locate the heavy atoms and to estimate the initial phases and the resulting map was then subjected to RESOLVE. The output of 455 non-H atoms, including 12 S atoms, one calcium ion and one chloride ion, were then subjected to ARP/wARP followed by REFMAC. With the improved phases, the automatic model building successfully built more than 85% of the 123 residues, excluding the N- and C-terminal residues. The final crystallographic R factor is 17.7% (R(free) = 21.7%). The refined model consists of 954 non-H protein atoms, 165 water O atoms, three 2-methyl-2,4-pentanediol (MPD) molecules, one calcium ion and one chloride ion. The present work is yet another example that shows the utility of single-wavelength anomalous scattering data for solving a protein structure.
Organizational Affiliation:
Bioinformatics Centre, Indian Institute of Science, Bangalore 560 012, India. sekar@physics.iisc.ernet.in