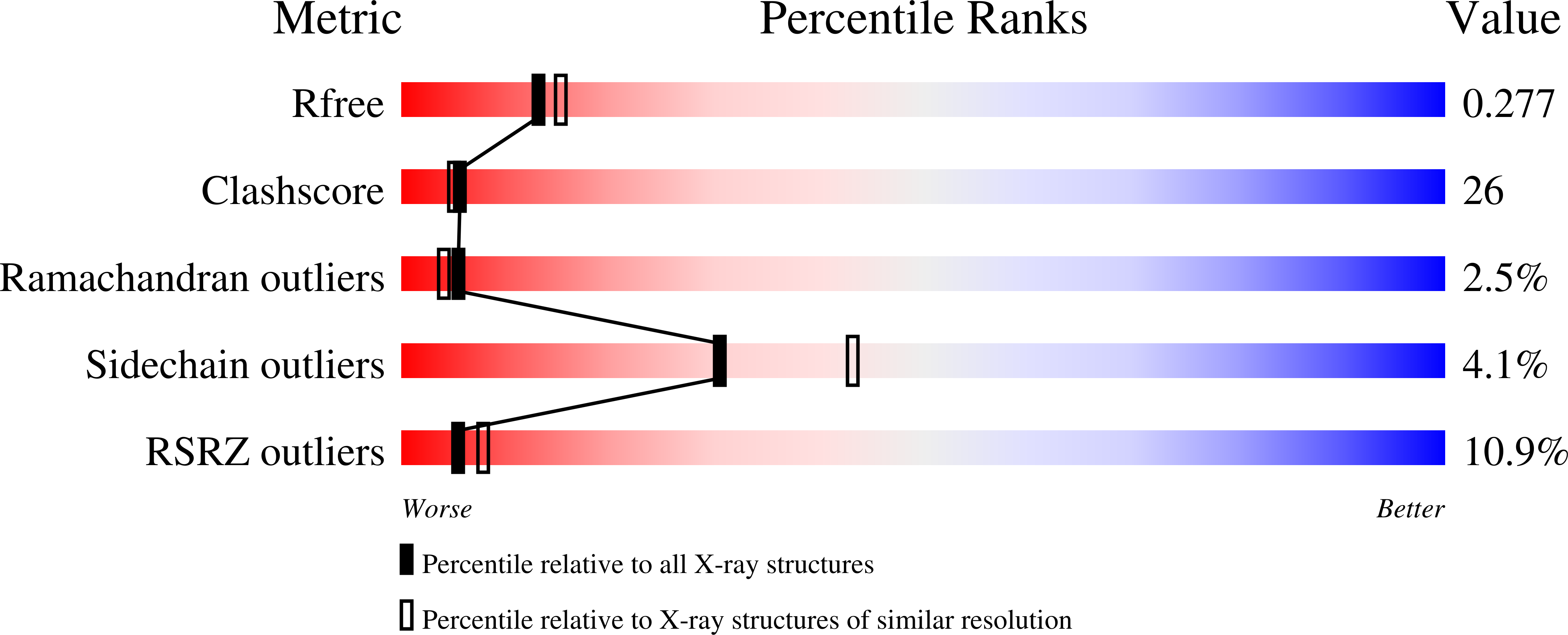
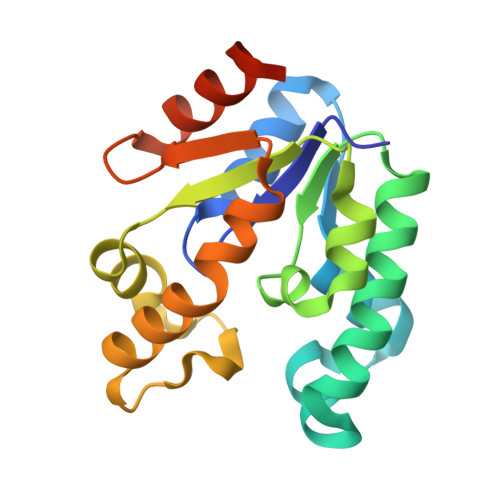
Structure of shikimate kinase from Mycobacterium tuberculosis reveals the binding of shikimic acid.
Pereira, J.H., de Oliveira, J.S., Canduri, F., Dias, M.V., Palma, M.S., Basso, L.A., Santos, D.S., de Azevedo, W.F.(2004) Acta Crystallogr D Biol Crystallogr 60: 2310-2319
- PubMed: 15583379
- DOI: https://doi.org/10.1107/S090744490402517X
- Primary Citation of Related Structures:
1WE2 - PubMed Abstract:
Tuberculosis made a resurgence in the mid-1980s and now kills approximately 3 million people a year. The re-emergence of tuberculosis as a public health threat, the high susceptibility of HIV-infected persons and the proliferation of multi-drug-resistant strains have created a need to develop new drugs. Shikimate kinase and other enzymes in the shikimate pathway are attractive targets for development of non-toxic antimicrobial agents, herbicides and anti-parasitic drugs, because the pathway is essential in these species whereas it is absent from mammals. The crystal structure of shikimate kinase from Mycobacterium tuberculosis (MtSK) complexed with MgADP and shikimic acid (shikimate) has been determined at 2.3 A resolution, clearly revealing the amino-acid residues involved in shikimate binding. This is the first three-dimensional structure of shikimate kinase complexed with shikimate. In MtSK, the Glu61 residue that is strictly conserved in shikimate kinases forms a hydrogen bond and salt bridge with Arg58 and assists in positioning the guanidinium group of Arg58 for shikimate binding. The carboxyl group of shikimate interacts with Arg58, Gly81 and Arg136 and the hydroxyl groups interact with Asp34 and Gly80. The crystal structure of MtSK-MgADP-shikimate will provide crucial information for the elucidation of the mechanism of the shikimate kinase-catalyzed reaction and for the development of a new generation of drugs against tuberculosis.
Organizational Affiliation:
Programa de Pós-Graduação em Biofísica Molecular, Departamento de Física, UNESP, São José do Rio Preto, SP 15054-000, Brazil.