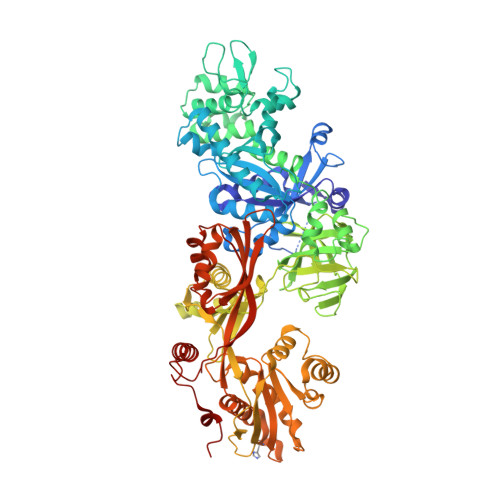
Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry.
Joergensen, R., Merrill, A.R., Yates, S.P., Marquez, V.E., Schwan, A.L., Boesen, T., Andersen, G.R.(2005) Nature 436: 979-984
- PubMed: 16107839
- DOI: https://doi.org/10.1038/nature03871
- Primary Citation of Related Structures:
1ZM2, 1ZM3, 1ZM4, 1ZM9 - PubMed Abstract:
The bacteria causing diphtheria, whooping cough, cholera and other diseases secrete mono-ADP-ribosylating toxins that modify intracellular proteins. Here, we describe four structures of a catalytically active complex between a fragment of Pseudomonas aeruginosa exotoxin A (ETA) and its protein substrate, translation elongation factor 2 (eEF2). The target residue in eEF2, diphthamide (a modified histidine), spans across a cleft and faces the two phosphates and a ribose of the non-hydrolysable NAD+ analogue, betaTAD. This suggests that the diphthamide is involved in triggering NAD+ cleavage and interacting with the proposed oxacarbenium intermediate during the nucleophilic substitution reaction, explaining the requirement of diphthamide for ADP ribosylation. Diphtheria toxin may recognize eEF2 in a manner similar to ETA. Notably, the toxin-bound betaTAD phosphates mimic the phosphate backbone of two nucleotides in a conformational switch of 18S rRNA, thereby achieving universal recognition of eEF2 by ETA.
- Centre for Structural Biology, Department of Molecular Biology, University of Aarhus, Gustav Wieds Vej 10C, DK-8000, Denmark.
Organizational Affiliation: