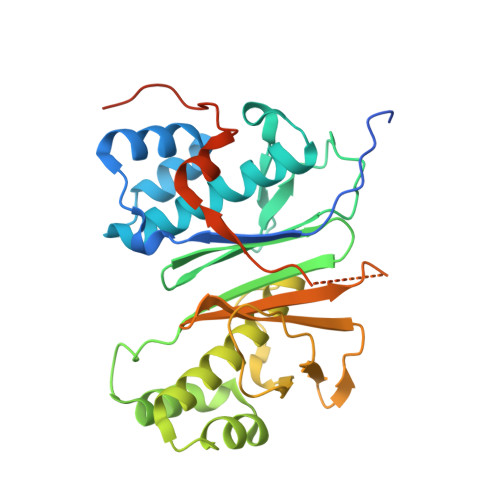
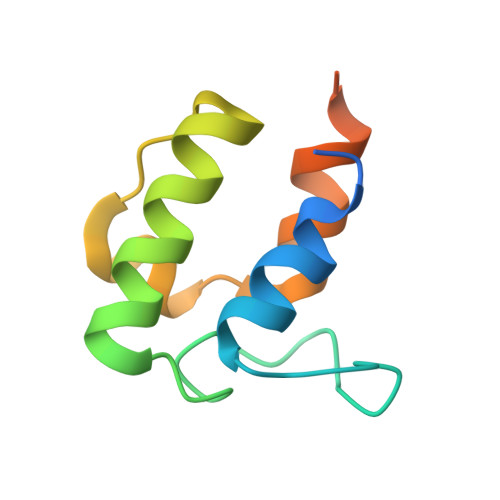
Mechanism and Substrate Recognition of Human Holo Acp Synthase.
Bunkoczi, G., Pasta, S., Joshi, A., Wu, X., Kavanagh, K.L., Smith, S., Oppermann, U.(2007) Chem Biol 14: 1243
- PubMed: 18022563
- DOI: https://doi.org/10.1016/j.chembiol.2007.10.013
- Primary Citation of Related Structures:
2BYD, 2C43, 2CG5 - PubMed Abstract:
Mammals utilize a single phosphopantetheinyl transferase for the posttranslational modification of at least three different apoproteins: the carrier protein components of cytosolic and mitochondrial fatty acid synthases and the aminoadipate semialdehyde reductase involved in lysine degradation. We determined the crystal structure of the human phosphopantetheinyl transferase, a eukaryotic phosphopantetheinyl transferase characterized, complexed with CoA and Mg(2+), and in ternary complex with CoA and ACP. The involvement of key residues in ligand binding and catalysis was confirmed by mutagenesis and kinetic analysis. Human phosphopantetheinyl transferase exhibits an alpha/beta fold and 2-fold pseudosymmetry similar to the Sfp phosphopantetheinyl transferase from Bacillus subtilis. Although the bound ACP exhibits a typical four-helix structure, its binding is unusual in that it is facilitated predominantly by hydrophobic interactions. A detailed mechanism is proposed describing the substrate binding and catalytic process.
- Structural Genomics Consortium, University of Oxford, Oxford OX3 7LD, UK.
Organizational Affiliation: