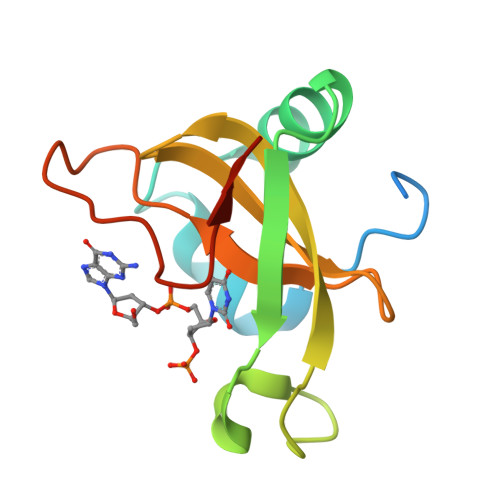
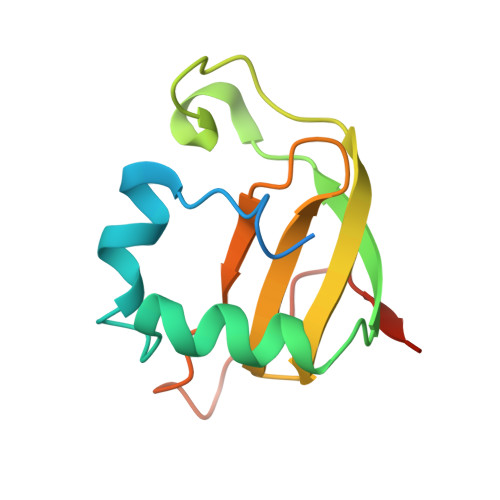
Structural basis for sequence-dependent recognition of colicin E5 tRNase by mimicking the mRNA-tRNA interaction
Yajima, S., Inoue, S., Ogawa, T., Nonaka, T., Ohsawa, K., Masaki, H.(2006) Nucleic Acids Res 34: 6074-6082
- PubMed: 17099236
- DOI: https://doi.org/10.1093/nar/gkl729
- Primary Citation of Related Structures:
2DFX, 2DJH - PubMed Abstract:
Colicin E5--a tRNase toxin--specifically cleaves QUN (Q: queuosine) anticodons of the Escherichia coli tRNAs for Tyr, His, Asn and Asp. Here, we report the crystal structure of the C-terminal ribonuclease domain (CRD) of E5 complexed with a substrate analog, namely, dGpdUp, at a resolution of 1.9 A. Thisstructure is the first to reveal the substrate recognition mechanism of sequence-specific ribonucleases. E5-CRD realized the strict recognition for both the guanine and uracil bases of dGpdUp forming Watson-Crick-type hydrogen bonds and ring stacking interactions, thus mimicking the codons of mRNAs to bind to tRNA anticodons. The docking model of E5-CRD with tRNA also suggests its substrate preference for tRNA over ssRNA. In addition, the structure of E5-CRD/dGpdUp along with the mutational analysis suggests that Arg33 may play an important role in the catalytic activity, and Lys25/Lys60 may also be involved without His in E5-CRD. Finally, the comparison of the structures of E5-CRD/dGpdUp and E5-CRD/ImmE5 (an inhibitor protein) complexes suggests that the binding mode of E5-CRD and ImmE5 mimics that of mRNA and tRNA; this may represent the evolutionary pathway of these proteins from the RNA-RNA interaction through the RNA-protein interaction of tRNA/E5-CRD.
Organizational Affiliation:
Department of Bioscience, Tokyo University of Agriculture, Sakuragaoka 1-1-1, Setagaya-ku, Tokyo 156-8502, Japan. yshun@nodai.ac.jp