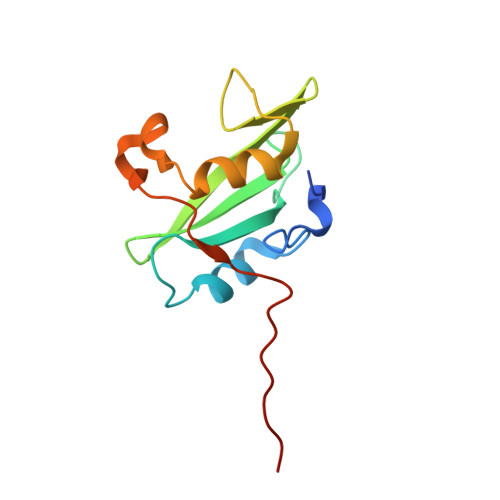
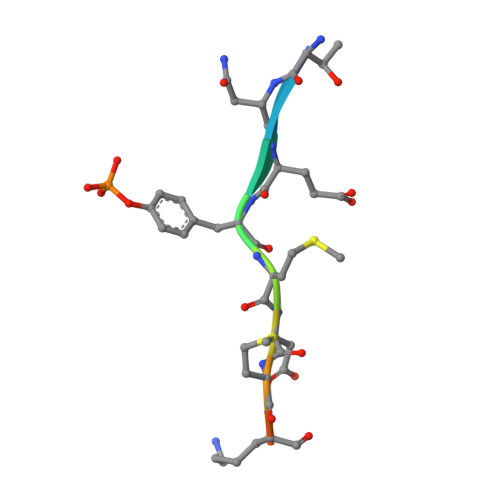
Crystal Structure of the Pi 3-Kinase P85 Amino-Terminal Sh2 Domain and its Phosphopeptide Complexes
Nolte, R.T., Eck, M.J., Schlessinger, J., Shoelson, S.E., Harrison, S.C.(1996) Nat Struct Biol 3: 364
- PubMed: 8599763
- DOI: https://doi.org/10.1038/nsb0496-364
- Primary Citation of Related Structures:
2IUG, 2IUH, 2IUI - PubMed Abstract:
Crystal structures of the amino-terminal SH2 domain of the p85alpha subunit of phosphatidylinositol (PI) 3-kinase, alone and in complex with phosphopeptides bearing pTyr-Met/Val-Xaa-Met motifs, show that phosphopeptides bind in the two-pronged manner seen in high-affinity Lck and Src SH2 complexes, with conserved interactions between the domain and the peptide segment from phosphotyrosine to Met+3. Peptide binding requires the rearrangement of a tyrosyl side chain in the BG loop to create the hydrophobic Met+3 binding pocket. The structures suggest a mechanism for the biological specificity exhibited by PI 3-kinase in its interactions with phosphoprotein partners.
- Howard Hughes Medical Institute and Laboratory of Molecular Medicine, Children's Hospital, Boston, Massachusetts 02115, USA.
Organizational Affiliation: