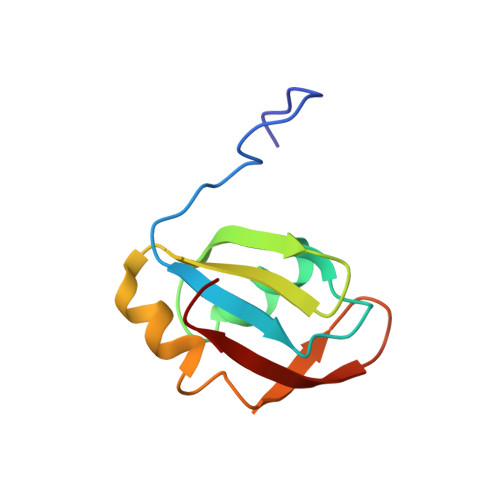
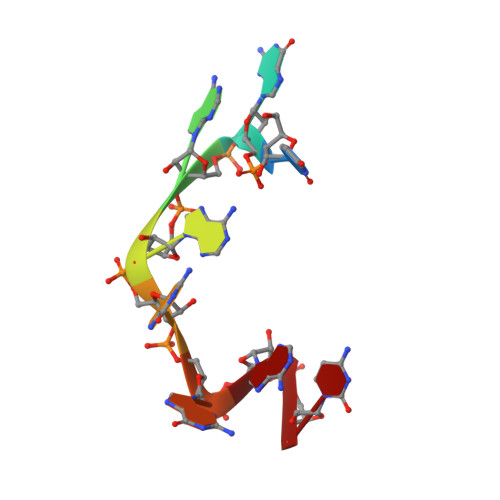Isolated pseudo-RNA-recognition motifs of SR proteins can regulate splicing using a noncanonical mode of RNA recognition.
Clery, A., Sinha, R., Anczukow, O., Corrionero, A., Moursy, A., Daubner, G.M., Valcarcel, J., Krainer, A.R., Allain, F.H.(2013) Proc Natl Acad Sci U S A 110: E2802-E2811
- PubMed: 23836656
- DOI: https://doi.org/10.1073/pnas.1303445110
- Primary Citation of Related Structures:
2M8D - PubMed Abstract:
Serine/arginine (SR) proteins, one of the major families of alternative-splicing regulators in Eukarya, have two types of RNA-recognition motifs (RRMs): a canonical RRM and a pseudo-RRM. Although pseudo-RRMs are crucial for activity of SR proteins, their mode of action was unknown. By solving the structure of the human SRSF1 pseudo-RRM bound to RNA, we discovered a very unusual and sequence-specific RNA-binding mode that is centered on one α-helix and does not involve the β-sheet surface, which typically mediates RNA binding by RRMs. Remarkably, this mode of binding is conserved in all pseudo-RRMs tested. Furthermore, the isolated pseudo-RRM is sufficient to regulate splicing of about half of the SRSF1 target genes tested, and the bound α-helix is a pivotal element for this function. Our results strongly suggest that SR proteins with a pseudo-RRM frequently regulate splicing by competing with, rather than recruiting, spliceosome components, using solely this unusual RRM.
- Institute for Molecular Biology and Biophysics, Swiss Federal Institute of Technology, 8093 Zurich, Switzerland.
Organizational Affiliation: