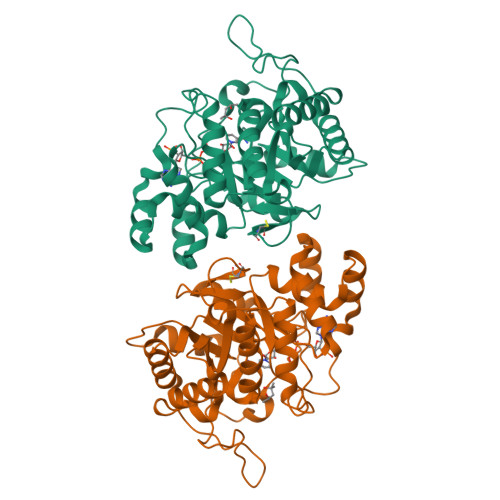
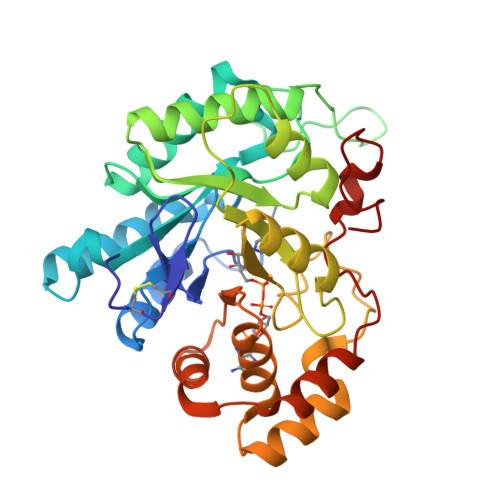
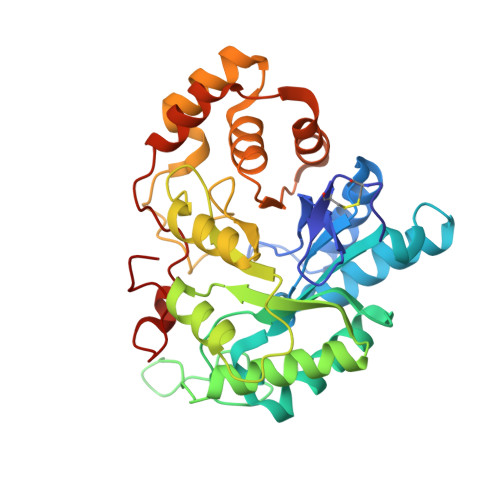
Structure of 3(17)alpha-hydroxysteroid dehydrogenase (AKR1C21) holoenzyme from an orthorhombic crystal form: an insight into the bifunctionality of the enzyme.
Dhagat, U., Carbone, V., Chung, R.P., Schulze-Briese, C., Endo, S., Hara, A., El-Kabbani, O.(2007) Acta Crystallogr Sect F Struct Biol Cryst Commun 63: 825-830
- PubMed: 17909281
- DOI: https://doi.org/10.1107/S1744309107040985
- Primary Citation of Related Structures:
2P5N - PubMed Abstract:
Mouse 3(17)alpha-hydroxysteroid dehydrogenase (AKR1C21) is a bifunctional enzyme that catalyses the oxidoreduction of the 3- and 17-hydroxy/keto groups of steroid substrates such as oestrogens, androgens and neurosteroids. The structure of the AKR1C21-NADPH binary complex was determined from an orthorhombic crystal belonging to space group P2(1)2(1)2(1) at a resolution of 1.8 A. In order to identify the factors responsible for the bifunctionality of AKR1C21, three steroid substrates including a 17-keto steroid, a 3-keto steroid and a 3alpha-hydroxysteroid were docked into the substrate-binding cavity. Models of the enzyme-coenzyme-substrate complexes suggest that Lys31, Gly225 and Gly226 are important for ligand recognition and orientation in the active site.
Organizational Affiliation:
Department of Medicinal Chemistry, Victorian College of Pharmacy, Monash University, Parkville, Victoria 3052, Australia.