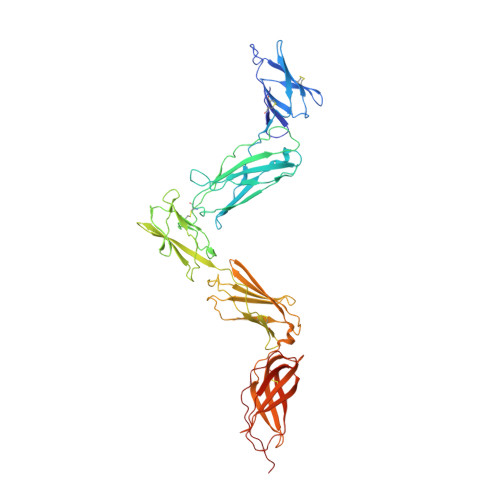
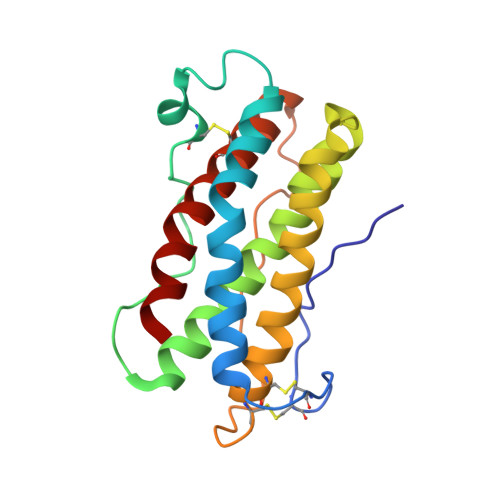
An unusual cytokine:Ig-domain interaction revealed in the crystal structure of leukemia inhibitory factor (LIF) in complex with the LIF receptor.
Huyton, T., Zhang, J.G., Luo, C.S., Lou, M.Z., Hilton, D.J., Nicola, N.A., Garrett, T.P.(2007) Proc Natl Acad Sci U S A 104: 12737-12742
- PubMed: 17652170
- DOI: https://doi.org/10.1073/pnas.0705577104
- Primary Citation of Related Structures:
2Q7N - PubMed Abstract:
Leukemia inhibitory factor (LIF) receptor is a cell surface receptor that mediates the actions of LIF and other IL-6 type cytokines through the formation of high-affinity signaling complexes with gp130. Here we present the crystal structure of a complex of mouse LIF receptor with human LIF at 4.0 A resolution. The structure is, to date, the largest cytokine receptor fragment determined by x-ray crystallography. The binding of LIF to its receptor via the central Ig-like domain is unlike other cytokine receptor complexes that bind ligand predominantly through their cytokine-binding modules. This structure, in combination with previous crystallographic studies, also provides a structural template to understand the formation and orientation of the high-affinity signaling complex between LIF, LIF receptor, and gp130.
- The Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, Victoria 3050, Australia.
Organizational Affiliation: