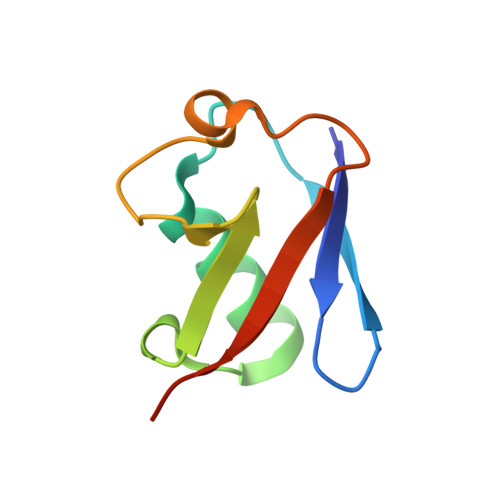
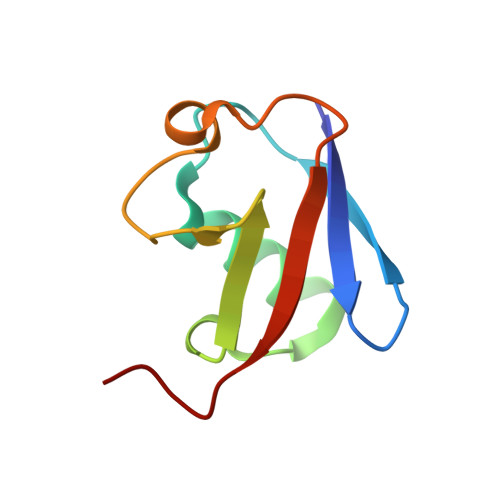
Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80
Sato, Y., Yoshikawa, A., Mimura, H., Yamashita, M., Yamagata, A., Fukai, S.(2009) EMBO J 28: 2461-2468
- PubMed: 19536136
- DOI: https://doi.org/10.1038/emboj.2009.160
- Primary Citation of Related Structures:
3A1Q - PubMed Abstract:
RAP80 has a key role in the recruitment of the Abraxas-BRCC36-BRCA1-BARD1 complex to DNA-damage foci for DNA repair through specific recognition of Lys 63-linked polyubiquitinated proteins by its tandem ubiquitin-interacting motifs (UIMs). Here, we report the crystal structure of the RAP80 tandem UIMs (RAP80-UIM1-UIM2) in complex with Lys 63-linked di-ubiquitin at 2.2 A resolution. The two UIMs, UIM1 and UIM2, and the alpha-helical inter-UIM region together form a continuous 60 A-long alpha-helix. UIM1 and UIM2 bind to the proximal and distal ubiquitin moieties, respectively. Both UIM1 and UIM2 of RAP80 recognize an Ile 44-centered hydrophobic patch on ubiquitin but neither UIM interacts with the Lys 63-linked isopeptide bond. Our structure suggests that the inter-UIM region forms a 12 A-long alpha-helix that ensures that the UIMs are arranged to enable specific binding of Lys 63-linked di-ubiquitin. This was confirmed by pull-down analyses using RAP80-UIM1-UIM2 mutants of various length inter-UIM regions. Further, we show that the Epsin1 tandem UIM, which has an inter-UIM region similar to that of RAP80-UIM1-UIM2, also selectively binds Lys 63-linked di-ubiquitin.
Organizational Affiliation:
Structural Biology Laboratory, Life Science Division, Synchrotron Radiation Research Organization and Institute of Molecular and Cellular Biosciences, The University of Tokyo, Tokyo, Japan.