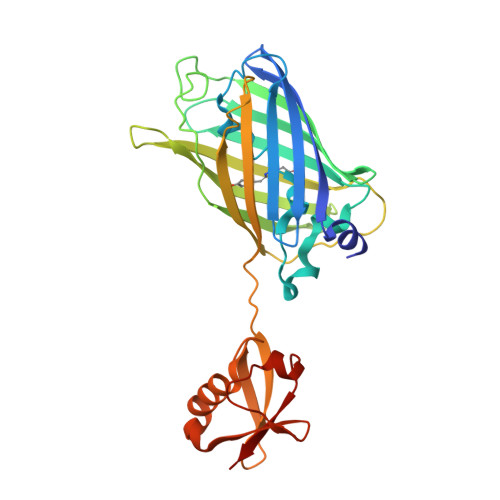
Crystallization of small proteins assisted by green fluorescent protein
Suzuki, N., Hiraki, M., Yamada, Y., Matsugaki, N., Igarashi, N., Kato, R., Dikic, I., Drew, D., Iwata, S., Wakatsuki, S., Kawasaki, M.(2010) Acta Crystallogr D Biol Crystallogr 66: 1059-1066
- PubMed: 20944239
- DOI: https://doi.org/10.1107/S0907444910032944
- Primary Citation of Related Structures:
3AI4, 3AI5 - PubMed Abstract:
The generation of crystal lattice contacts by proteinaceous tags fused to target proteins is an attractive approach to aid in the crystallization of otherwise intractable proteins. Here, the use of green fluorescent protein (GFP) fusions for this purpose is demonstrated, using ubiquitin and the ubiquitin-binding motif (UBM) of Y-family polymerase ι as examples. The structure of the GFP-ubiquitin fusion protein revealed that the crystal lattice was formed by GFP moieties. Ubiquitin was accommodated in the lattice through interactions with the peripheral loops of GFP. However, in the GFP-UBM fusion crystal UBM formed extensive interactions with GFP and these interactions, together with UBM dimerization, mediated the crystal packing. Interestingly, the tyrosine residues that are involved in mediating crystal contacts in both GFP-ubiquitin and GFP-UBM crystals are arranged in a belt on the surface of the β-barrel structure of GFP. Therefore, it is likely that GFP can assist in the crystallization of small proteins and of protein domains in general.
Organizational Affiliation:
Structural Biology Research Center, Photon Factory, Institute of Materials Structure Science, High Energy Accelerator Research Organization (KEK), Tsukuba, Ibaraki 305-0801, Japan.