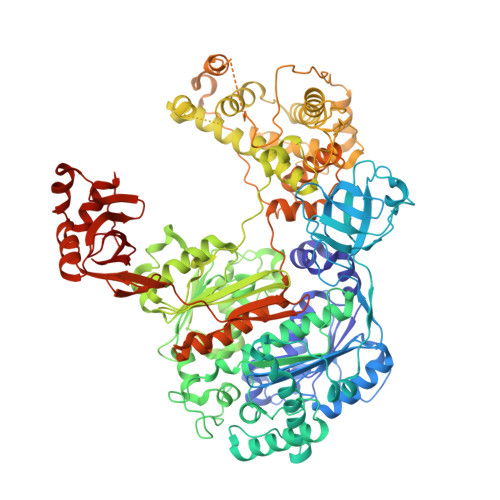
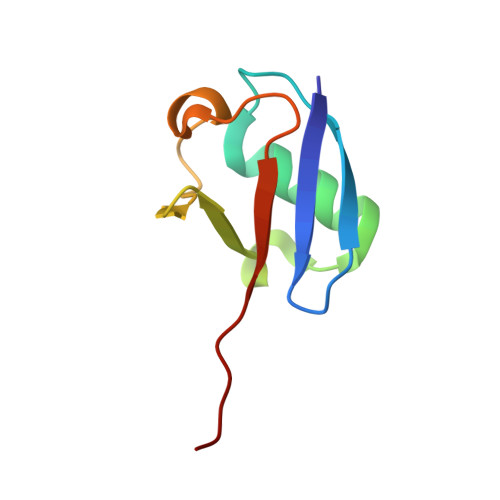
Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes.
Lee, I., Schindelin, H.(2008) Cell 134: 268-278
- PubMed: 18662542
- DOI: https://doi.org/10.1016/j.cell.2008.05.046
- Primary Citation of Related Structures:
3CMM - PubMed Abstract:
Ubiquitin (Ub) and ubiquitin-like proteins (Ubls) are conjugated to their targets by specific cascades involving three classes of enzymes, E1, E2, and E3. Each E1 adenylates the C terminus of its cognate Ubl, forms a E1 approximately Ubl thioester intermediate, and ultimately generates a thioester-linked E2 approximately Ubl product. We have determined the crystal structure of yeast Uba1, revealing a modular architecture with individual domains primarily mediating these specific activities. The negatively charged C-terminal ubiquitin-fold domain (UFD) is primed for binding of E2s and recognizes their positively charged first alpha helix via electrostatic interactions. In addition, a mobile loop from the domain harboring the E1 catalytic cysteine contributes to E2 binding. Significant, experimentally observed motions in the UFD around a hinge in the linker connecting this domain to the rest of the enzyme suggest a conformation-dependent mechanism for the transthioesterification function of Uba1; however, this mechanism clearly differs from that of other E1 enzymes.
- Department of Biochemistry and Cell Biology, Stony Brook University, Stony Brook, NY 11794-5215, USA.
Organizational Affiliation: