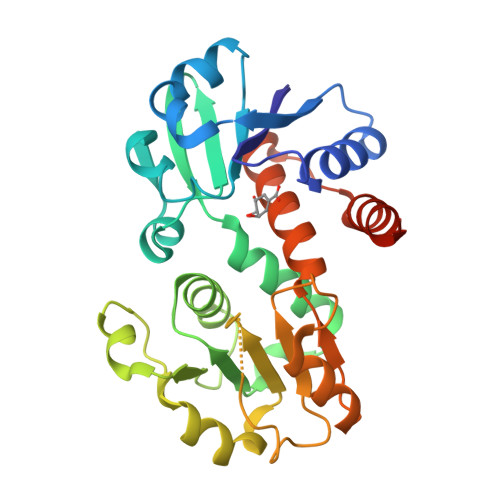
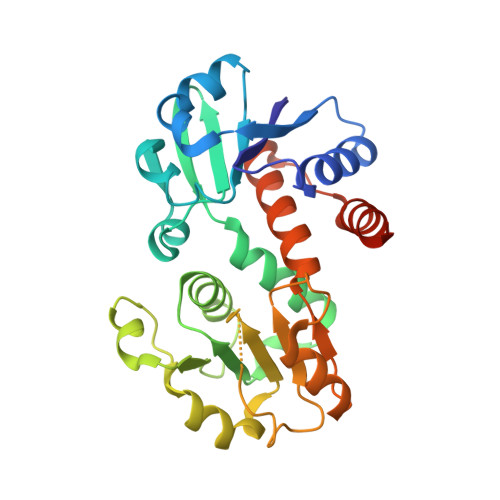
X-ray crystallographic and enzymatic analyses of shikimate dehydrogenase from Staphylococcus epidermidis
Han, C., Hu, T., Wu, D., Qu, S., Zhou, J., Ding, J., Shen, X., Qu, D., Jiang, H.(2009) FEBS J 276: 1125-1139
- PubMed: 19215302
- DOI: https://doi.org/10.1111/j.1742-4658.2008.06856.x
- Primary Citation of Related Structures:
3DON, 3DOO - PubMed Abstract:
Shikimate dehydrogenase (SDH) catalyzes the NADPH-dependent reduction of 3-dehydroshikimate to shikimate in the shikimate pathway. In this study, we determined the kinetic properties and crystal structures of Staphylococcus epidermidis SDH (SeSDH) both in its ligand-free form and in complex with shikimate. SeSDH has a k(cat) of 22.8 s(-1) and a K(m) of 73 mum towards shikimate, and a K(m) of 100 microM towards NADP. The overall folding of SeSDH comprises the N-terminal alpha/beta domain for substrate binding and the C-terminal Rossmann fold for NADP binding. The active site is within a large groove between the two domains. Residue Tyr211, normally regarded as important for substrate binding, does not interact with shikimate in the binary SeSDH-shikimate complex structure. However, the Y211F mutation leads to a significant decrease in k(cat) and a minor increase in the K(m) for shikimate. The results indicate that the main function of Tyr211 may be to stabilize the catalytic intermediate during catalysis. The NADP-binding domain of SeSDH is less conserved. The usually long helix specifically recognizing the adenine ribose phosphate is substituted with a short 3(10) helix in the NADP-binding domain. Moreover, the interdomain angle of SeSDH is the widest among all known SDH structures, indicating an inactive 'open' state of the SeSDH structure. Thus, a 'closing' process might occur upon NADP binding to bring the cofactor close to the substrate for catalysis.
Organizational Affiliation:
Institutes of Biomedical Sciences and Key Laboratory of Medical Molecular Virology, Institute of Medical Microbiology, Shanghai Medical College, Fudan University, China.