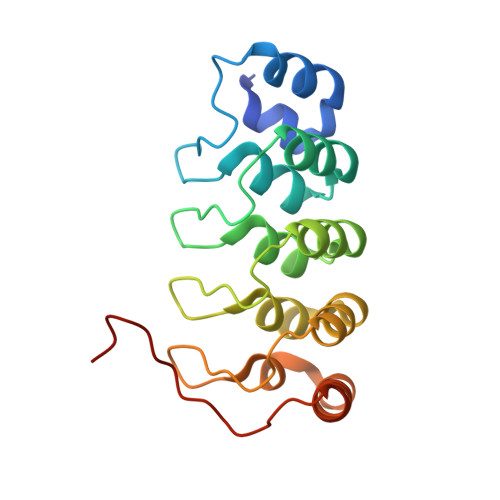
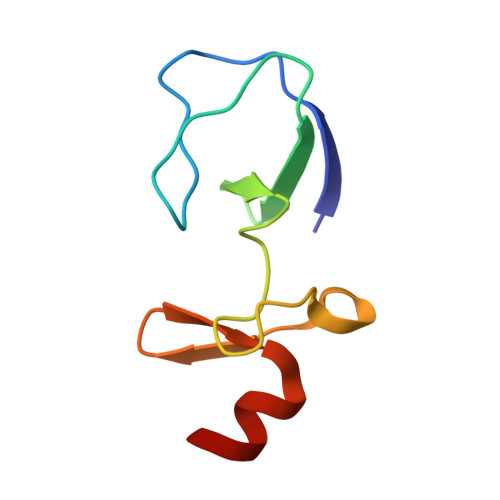
The structural basis of integrin-linked kinase-PINCH interactions.
Chiswell, B.P., Zhang, R., Murphy, J.W., Boggon, T.J., Calderwood, D.A.(2008) Proc Natl Acad Sci U S A 105: 20677-20682
- PubMed: 19074270
- DOI: https://doi.org/10.1073/pnas.0811415106
- Primary Citation of Related Structures:
3F6Q - PubMed Abstract:
The heterotrimeric complex between integrin-linked kinase (ILK), PINCH, and parvin is an essential signaling platform, serving as a convergence point for integrin and growth-factor signaling and regulating cell adhesion, spreading, and migration. We report a 1.6-A crystal structure of the ILK ankyrin repeat domain bound to the PINCH1 LIM1 domain, revealing the molecular basis of ILK-PINCH interactions and providing a structural description of this region of ILK. This structure identifies 5 ankyrin repeats in ILK, explains previous deletion mutagenesis data, permits identification of ILK and PINCH1 point mutations that disrupt the interaction, shows how zincs are coordinated by PINCH1 LIM1, and suggests that conformational flexibility and twisting between the 2 zinc fingers within the LIM1 domain may be important for ILK binding. These data provide an atomic-resolution description of a key interaction in the ILK-PINCH-parvin scaffolding complex.
- Department of Pharmacology, Yale Cancer Center and Interdepartmental Program in Vascular Biology and Transplantation, Yale University School of Medicine, 333 Cedar Street, New Haven, CT 06520, USA.
Organizational Affiliation: