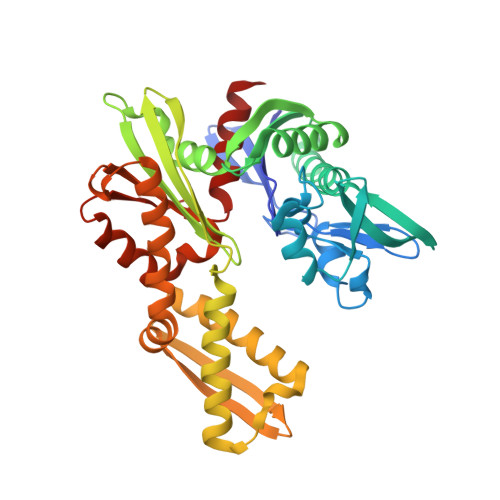
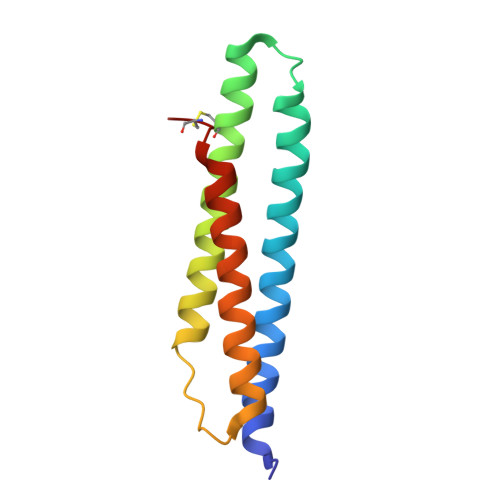
Novel adenosine-derived inhibitors of 70 kDa heat shock protein, discovered through structure-based design
Williamson, D.S., Borgognoni, J., Clay, A., Daniels, Z., Dokurno, P., Drysdale, M.J., Foloppe, N., Francis, G.L., Graham, C.J., Howes, R., Macias, A.T., Murray, J.B., Parsons, R., Shaw, T., Surgenor, A.E., Terry, L., Wang, Y., Wood, M., Massey, A.J.(2009) J Med Chem 52: 1510-1513
- PubMed: 19256508
- DOI: https://doi.org/10.1021/jm801627a
- Primary Citation of Related Structures:
3FZF, 3FZH, 3FZK, 3FZL, 3FZM - PubMed Abstract:
The design and synthesis of novel adenosine-derived inhibitors of HSP70, guided by modeling and X-ray crystallographic structures of these compounds in complex with HSC70/BAG-1, is described. Examples exhibited submicromolar affinity for HSP70, were highly selective over HSP90, and some displayed potency against HCT116 cells. Exposure of compound 12 to HCT116 cells caused significant reduction in cellular levels of Raf-1 and Her2 at concentrations similar to that which caused cell growth arrest.
- Vernalis (R&D) Ltd., Granta Park, Great Abington, Cambridge CB21 6GB, United Kingdom. d.williamson@vernalis.com
Organizational Affiliation: