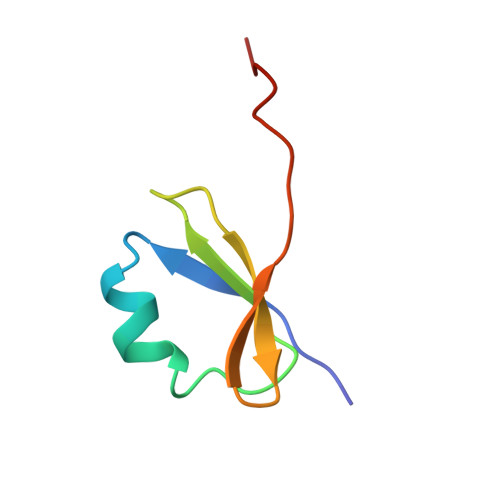
Crystal structure of human collagen XVIII trimerization domain: A novel collagen trimerization Fold.
Boudko, S.P., Sasaki, T., Engel, J., Lerch, T.F., Nix, J., Chapman, M.S., Bachinger, H.P.(2009) J Mol Biology 392: 787-802
- PubMed: 19631658
- DOI: https://doi.org/10.1016/j.jmb.2009.07.057
- Primary Citation of Related Structures:
3HON, 3HSH - PubMed Abstract:
Collagens contain a unique triple-helical structure with a repeating sequence -G-X-Y-, where proline and hydroxyproline are major constituents in X and Y positions, respectively. Folding of the collagen triple helix requires trimerization domains. Once trimerized, collagen chains are correctly aligned and the folding of the triple helix proceeds in a zipper-like fashion. Here we report the isolation, characterization, and crystal structure of the trimerization domain of human type XVIII collagen, a member of the multiplexin family. This domain differs from all other known trimerization domains in other collagens and exhibits a high trimerization potential at picomolar concentrations. Strong chain association and high specificity of binding are needed for multiplexins, which are present at very low levels.
- Research Department of Shriners Hospital for Children, Portland, OR 97239, USA.
Organizational Affiliation: