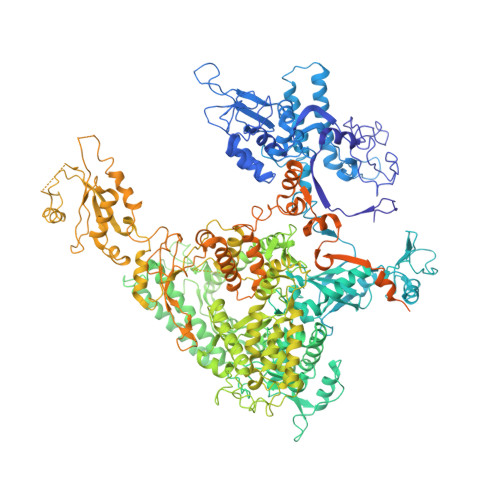
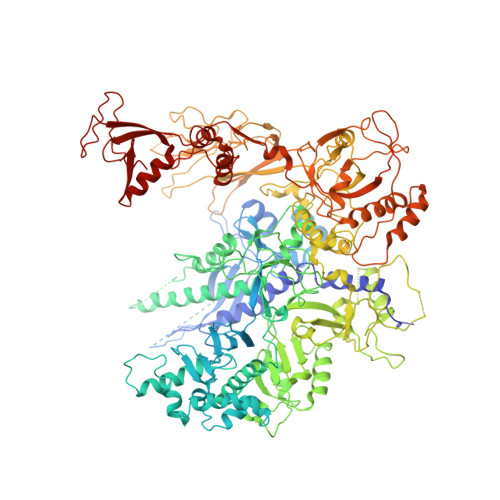
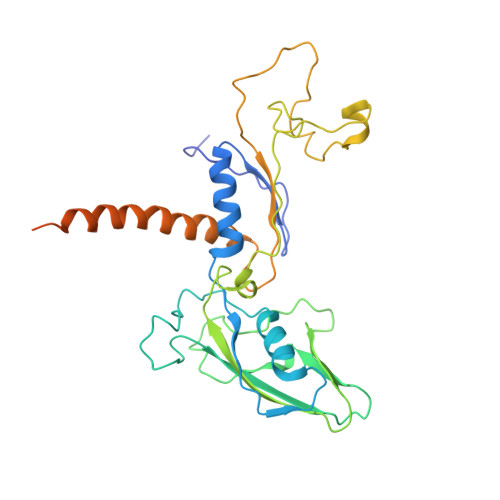
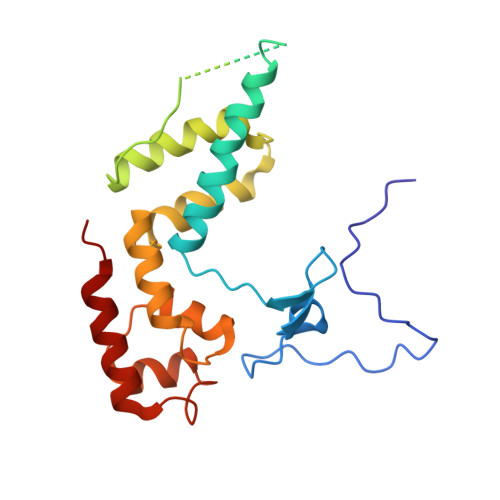
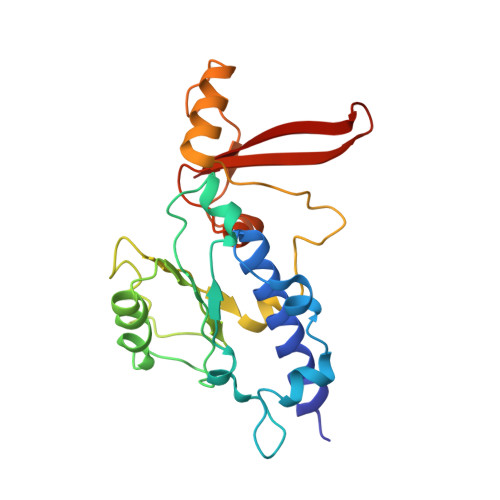
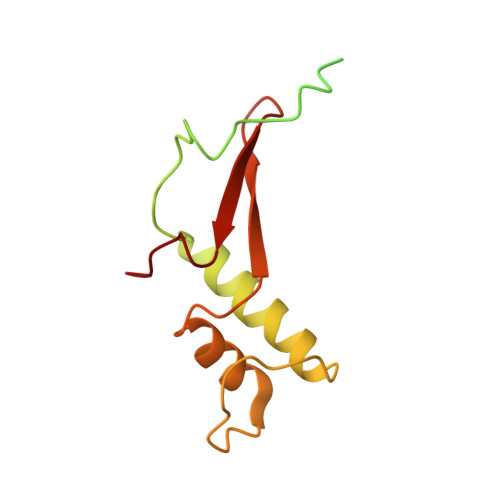
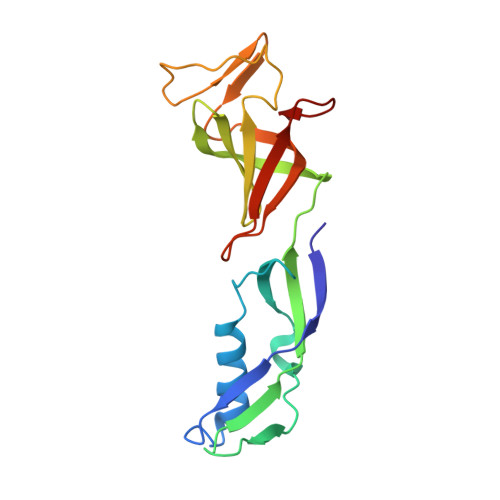
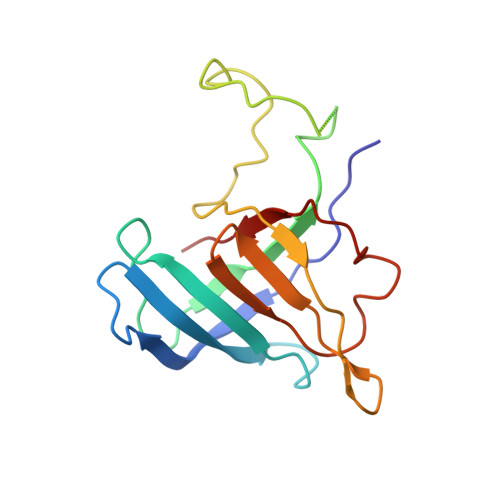
Molecular basis of transcriptional mutagenesis at 8-oxoguanine
Damsma, G.E., Cramer, P.(2009) J Biol Chem 284: 31658-31663
- PubMed: 19758983
- DOI: https://doi.org/10.1074/jbc.M109.022764
- Primary Citation of Related Structures:
3I4M, 3I4N - PubMed Abstract:
Structure-function analysis has revealed the mechanism of yeast RNA polymerase II transcription at 8-oxoguanine (8-oxoG), the major DNA lesion resulting from oxidative stress. When polymerase II encounters 8-oxoG in the DNA template strand, it can misincorporate adenine, which forms a Hoogsteen bp with 8-oxoG at the active center. This requires rotation of the 8-oxoG base from the standard anti- to an uncommon syn-conformation, which likely occurs during 8-oxoG loading into the active site. The misincorporated adenine escapes intrinsic proofreading, resulting in transcriptional mutagenesis that is observed directly by mass spectrometric RNA analysis.
Organizational Affiliation:
Gene Center and Center for Integrated Protein Science Munich (CIPSM), Department of Chemistry and Biochemistry, Ludwig-Maximilians-Universität München, Feodor-Lynen-Strasse 25, 81377 Munich, Germany.