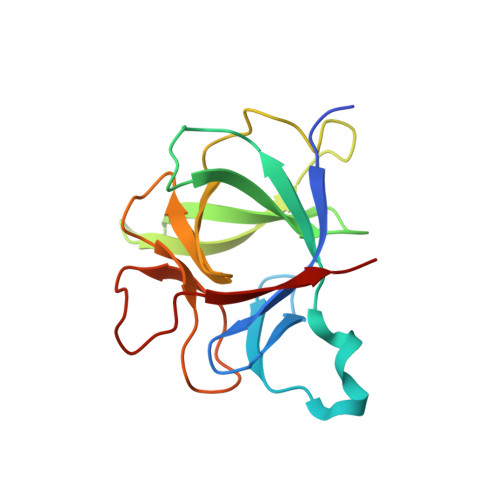
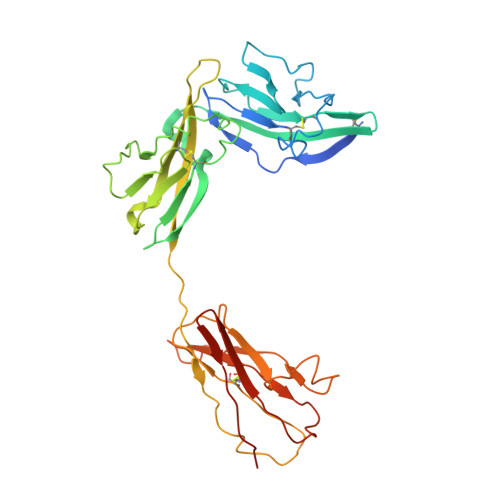
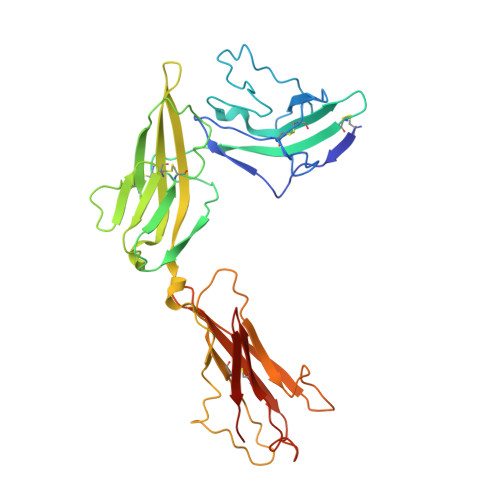
Structural insights into the assembly and activation of IL-1beta with its receptors
Wang, D.L., Zhang, S.Y., Li, L., Liu, X., Mei, K.R., Wang, X.Q.(2010) Nat Immunol 11: 905-911
- PubMed: 20802483
- DOI: https://doi.org/10.1038/ni.1925
- Primary Citation of Related Structures:
3O4O - PubMed Abstract:
Interleukin 1β (IL-1β) is a key orchestrator of inflammation and host defense that exerts its effects through IL-1 receptor type I (IL-1RI) and IL-1 receptor accessory protein (IL-1RAcP). How IL-1RAcP is recruited by IL-1β-IL-1RI to form the signaling-competent complex remains elusive. Here we present the crystal structure of IL-1β bound to IL-1 receptor type II (IL-1RII) and IL-1RAcP. IL-1β-IL-1RII generated a composite binding surface to recruit IL-1RAcP. Biochemical analysis demonstrated that IL-1β-IL-1RI and IL-1β-IL-1RII interacted similarly with IL-1RAcP. It also showed the importance of two loops of IL-1 receptor antagonist (IL-1Ra) in determining its antagonism. Our results provide a structural basis for assembly and activation of the IL-1 receptor and offer a general cytokine-receptor architecture that governs the IL-1 family of cytokines.
- Center for Structural Biology, School of Life Sciences, Ministry of Education Key Laboratory of Bioinformatics, Tsinghua University, Beijing, China.
Organizational Affiliation: