Discovery of novel and orally active FXR agonists for the potential treatment of dyslipidemia & diabetes
Richter, H.G.F., Benson, G.M., Blum, D., Chaput, E., Feng, S., Gardes, C., Grether, U., Hartman, P., Kuhn, B., Martin, R.E., Plancher, J.-M., Rudolph, M.G., Schuler, F., Taylor, S., Bleicher, K.H.(2010) Bioorg Med Chem Lett 21: 191-194
- PubMed: 21134747
- DOI: https://doi.org/10.1016/j.bmcl.2010.11.039
- Primary Citation of Related Structures:
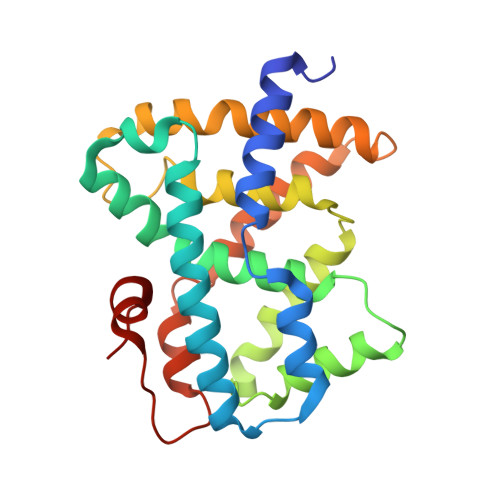
3OKH, 3OKI - PubMed Abstract:
Herein we describe the synthesis and structure activity relationship of a new class of FXR agonists identified from a high-throughput screening campaign. Further optimization of the original hits led to molecules that were highly active in an LDL-receptor KO model for dyslipidemia. The most promising candidate is discussed in more detail.
- F Hoffmann-La Roche AG, Grenzacherstrasse, CH-4070 Basel, Switzerland.
Organizational Affiliation: