Structure of a highly mutated, thermostable \xCE\xB1-amylase variant
Hein, K.L., Ganshaw, G., Bott, R., Nissen, P.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Alpha amylase | 435 | Pyrococcus woesei | Mutation(s): 0 EC: 3.2.1.1 | 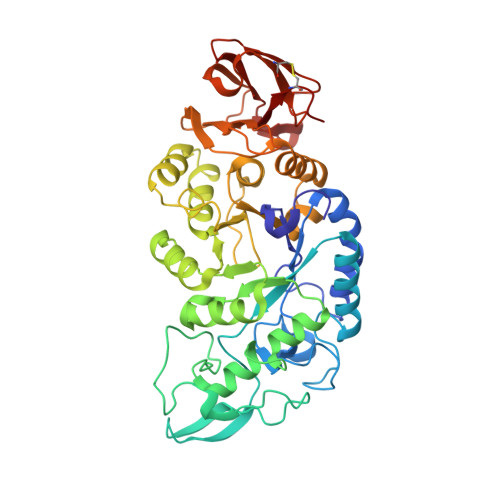 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| TRS Query on TRS | J [auth A] | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL C4 H12 N O3 LENZDBCJOHFCAS-UHFFFAOYSA-O |  | ||
| SO4 Query on SO4 | K [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| ZN Query on ZN | E [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| CA Query on CA | F [auth A], G [auth A], H [auth A], I [auth A] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_900012 Query on PRD_900012 | B | beta-cyclodextrin | Oligosaccharide / Drug delivery |  | |
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_900003 Query on PRD_900003 | C, D | sucrose | Oligosaccharide / Nutrient |  | |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 59.9 | α = 90 |
| b = 59.9 | β = 90 |
| c = 272.41 | γ = 90 |
| Software Name | Purpose |
|---|---|
| Blu-Ice | data collection |
| PHASER | phasing |
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |