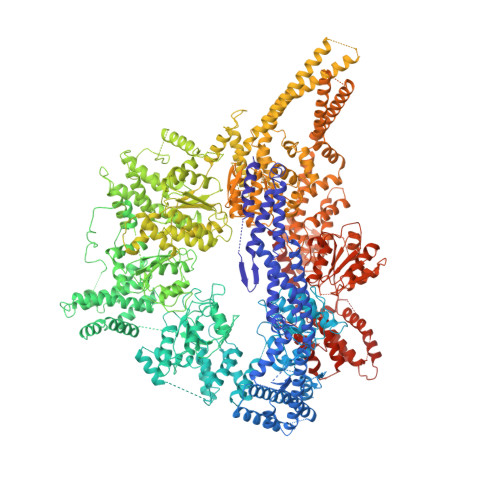
Crystal structure of the dynein motor domain.
Carter, A.P., Cho, C., Jin, L., Vale, R.D.(2011) Science 331: 1159-1165
- PubMed: 21330489
- DOI: https://doi.org/10.1126/science.1202393
- Primary Citation of Related Structures:
3QMZ - PubMed Abstract:
Dyneins are microtubule-based motor proteins that power ciliary beating, transport intracellular cargos, and help to construct the mitotic spindle. Evolved from ring-shaped hexameric AAA-family adenosine triphosphatases (ATPases), dynein's large size and complexity have posed challenges for understanding its structure and mechanism. Here, we present a 6 angstrom crystal structure of a functional dimer of two ~300-kilodalton motor domains of yeast cytoplasmic dynein. The structure reveals an unusual asymmetric arrangement of ATPase domains in the ring-shaped motor domain, the manner in which the mechanical element interacts with the ATPase ring, and an unexpected interaction between two coiled coils that create a base for the microtubule binding domain. The arrangement of these elements provides clues as to how adenosine triphosphate-driven conformational changes might be transmitted across the motor domain.
Organizational Affiliation:
Department of Cellular and Molecular Pharmacology, Howard Hughes Medical Institute, University of California-San Francisco, 600 16th Street, San Francisco, CA 94158, USA. cartera@mrc-lmb.cam.ac.uk