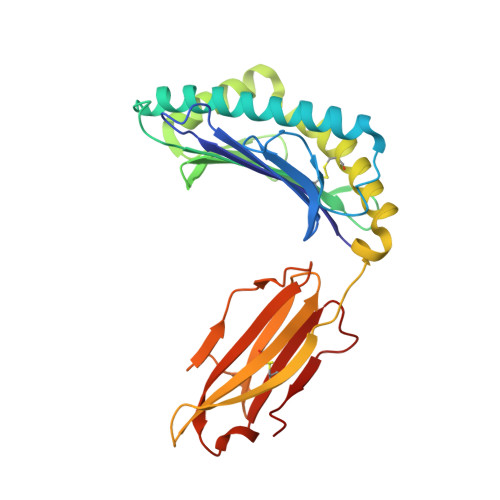
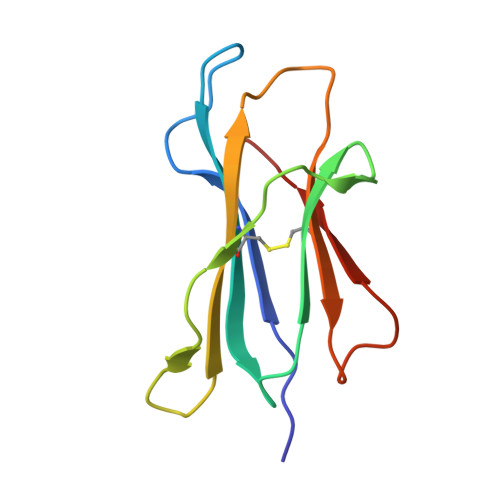
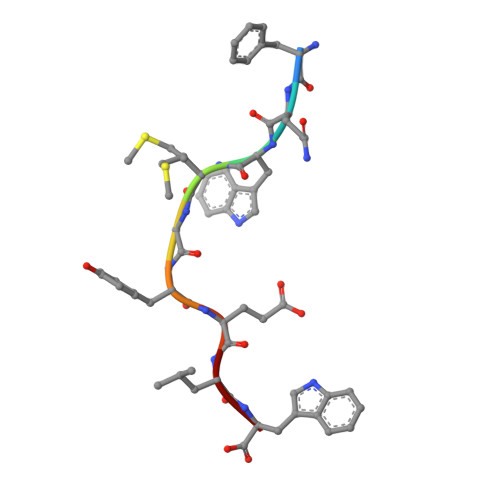
Structural basis of diverse peptide accommodation by the rhesus macaque MHC class I molecule Mamu-B*17: insights into immune protection from simian immunodeficiency virus
Wu, Y., Gao, F., Liu, J., Qi, J.X., Gostick, E., Price, D.A., Gao, G.F.(2011) J Immunol 187: 6382-6392
- PubMed: 22084443
- DOI: https://doi.org/10.4049/jimmunol.1101726
- Primary Citation of Related Structures:
3RWC, 3RWD, 3RWE, 3RWF, 3RWG, 3RWH, 3RWI, 3RWJ - PubMed Abstract:
The MHC class I molecule Mamu-B*17 has been associated with elite control of SIV infection in rhesus macaques, akin to the protective effects described for HLA-B*57 in HIV-infected individuals. In this study, we determined the crystal structures of Mamu-B*17 in complex with eight different peptides corresponding to immunodominant SIV(mac)239-derived CD8(+) T cell epitopes: HW8 (HLEVQGYW), GW10 (GSHLEVQGYW), MW9 (MHPAQTSQW), QW9 (QTSQWDDPW), FW9 (FQWMGYELW), MF8 (MRHVLEPF), IW9 (IRYPKTFGW), and IW11 (IRYPKTFGWLW). The structures reveal that not only P2, but also P1 and P3, can be used as N-terminal anchor residues by Mamu-B*17-restricted peptides. Moreover, the N-terminal anchor residues exhibit a broad chemical specificity, encompassing basic (H and R), bulky polar aliphatic (Q), and small (T) residues. In contrast, Mamu-B*17 exhibits a very narrow preference for aromatic residues (W and F) at the C terminus, similar to that displayed by HLA-B*57. Flexibility within the whole peptide-binding groove contributes to the accommodation of these diverse peptides, which adopt distinct conformations. Furthermore, the unusually large pocket D enables compensation from other peptide residues if P3 is occupied by an amino acid with a small side chain. In addition, residues located at likely TCR contact regions present highly flexible conformations, which may impact TCR repertoire profiles. These findings provide novel insights into the structural basis of diverse peptide accommodation by Mamu-B*17 and highlight unique atomic features that might contribute to the protective effect of this MHC I molecule in SIV-infected rhesus macaques.
- CAS Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, The People's Republic of China.
Organizational Affiliation: