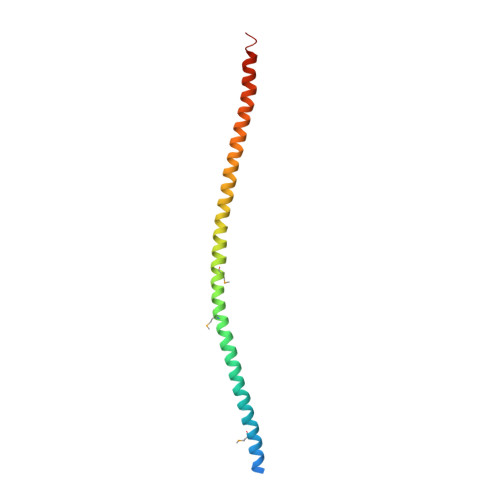The structure of vimentin linker 1 and rod 1B domains characterized by site-directed spin-labeling electron paramagnetic resonance (SDSL-EPR) and X-ray crystallography.
Aziz, A., Hess, J.F., Budamagunta, M.S., Voss, J.C., Kuzin, A.P., Huang, Y.J., Xiao, R., Montelione, G.T., FitzGerald, P.G., Hunt, J.F.(2012) J Biological Chem 287: 28349-28361
- PubMed: 22740688
- DOI: https://doi.org/10.1074/jbc.M111.334011
- Primary Citation of Related Structures:
3UF1 - PubMed Abstract:
Despite the passage of ∼30 years since the complete primary sequence of the intermediate filament (IF) protein vimentin was reported, the structure remains unknown for both an individual protomer and the assembled filament. In this report, we present data describing the structure of vimentin linker 1 (L1) and rod 1B. Electron paramagnetic resonance spectra collected from samples bearing site-directed spin labels demonstrate that L1 is not a flexible segment between coiled-coils (CCs) but instead forms a rigid, tightly packed structure. An x-ray crystal structure of a construct containing L1 and rod 1B shows that it forms a tetramer comprising two equivalent parallel CC dimers that interact with one another in the form of a symmetrical anti-parallel dimer. Remarkably, the parallel CC dimers are themselves asymmetrical, which enables them to tetramerize rather than undergoing higher order oligomerization. This functionally vital asymmetry in the CC structure, encoded in the primary sequence of rod 1B, provides a striking example of evolutionary exploitation of the structural plasticity of proteins. EPR and crystallographic data consistently suggest that a very short region within L1 represents a minor local distortion in what is likely to be a continuous CC from the end of rod 1A through the entirety of rod 1B. The concordance of this structural model with previously published cross-linking and spectral data supports the conclusion that the crystallographic oligomer represents a native biological structure.
- Department of Cell Biology and Human Anatomy, University of California, Davis, California 95616, USA.
Organizational Affiliation: