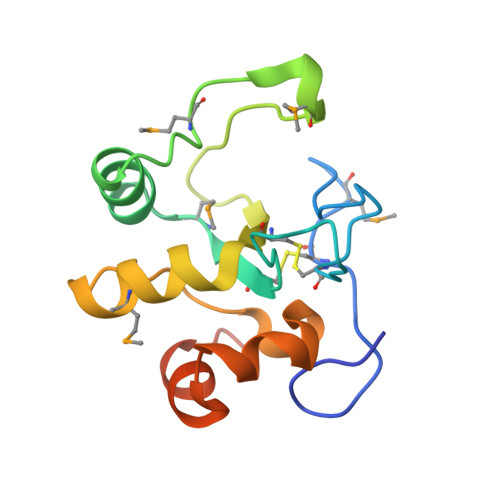
Crystal Structure of the Q2S0R5 protein from Salinibacter ruber, Northeast Structural Genomics Consortium Target SrR207
Vorobiev, S., Su, M., Seetharaman, J., Maglaqui, M., Xiao, R., Kohan, E., Wang, D., Everett, J.K., Acton, T.B., Montelione, G.T., Tong, L., Hunt, J.F.To be published.